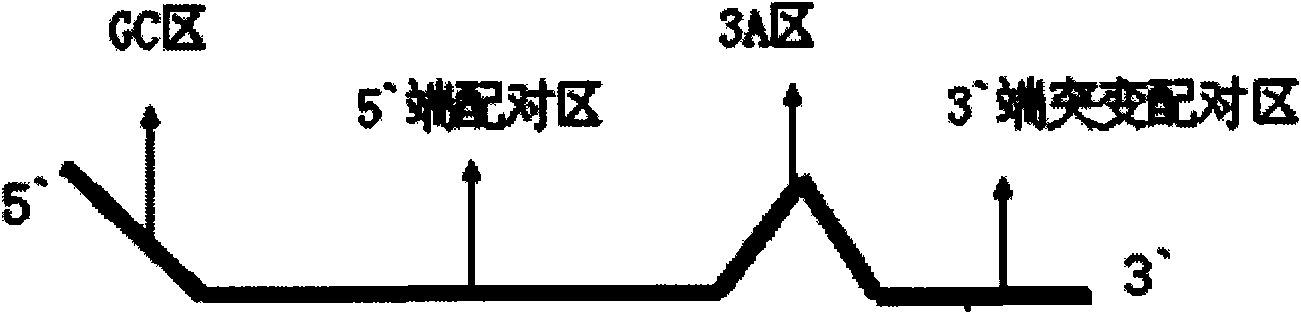
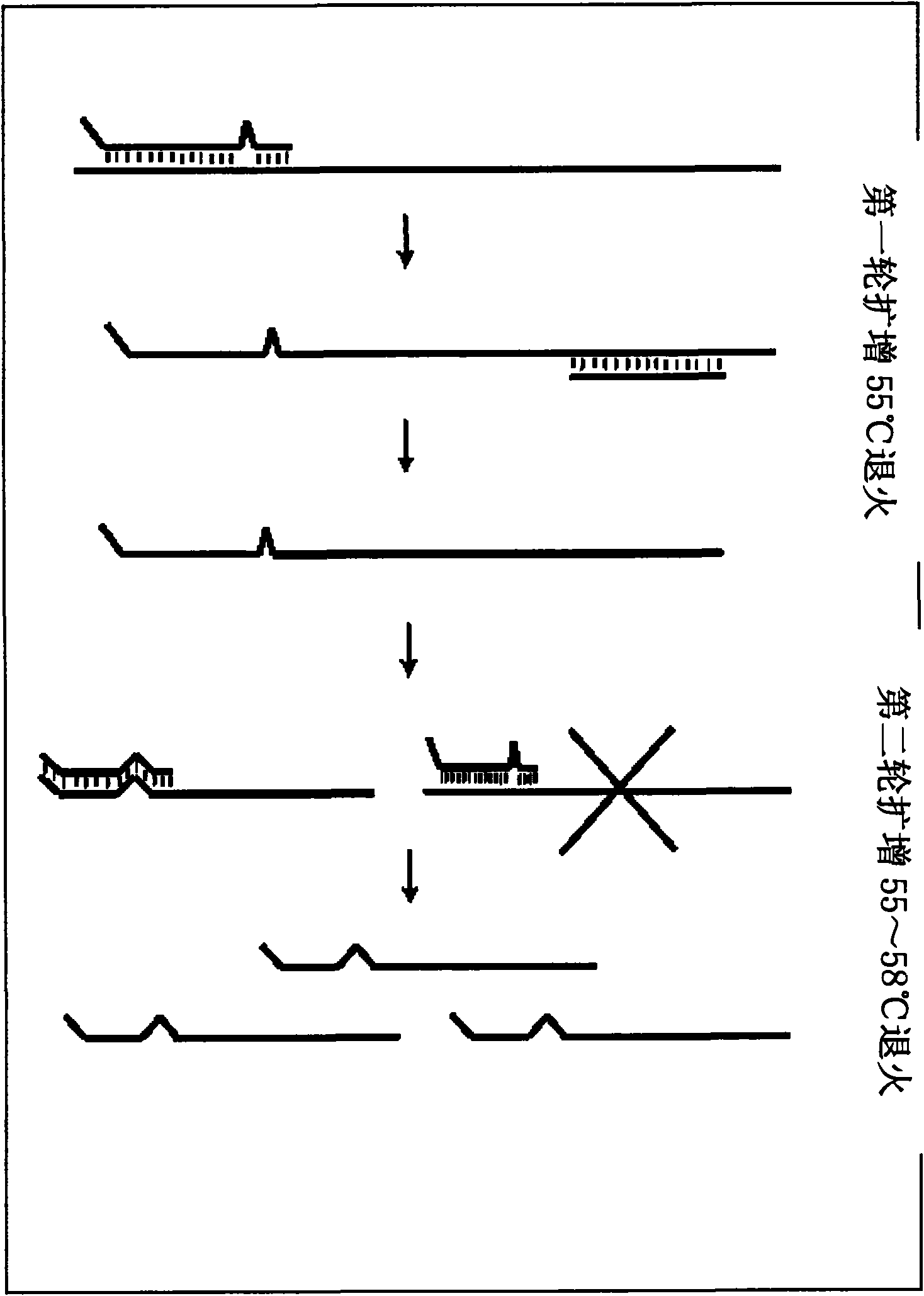
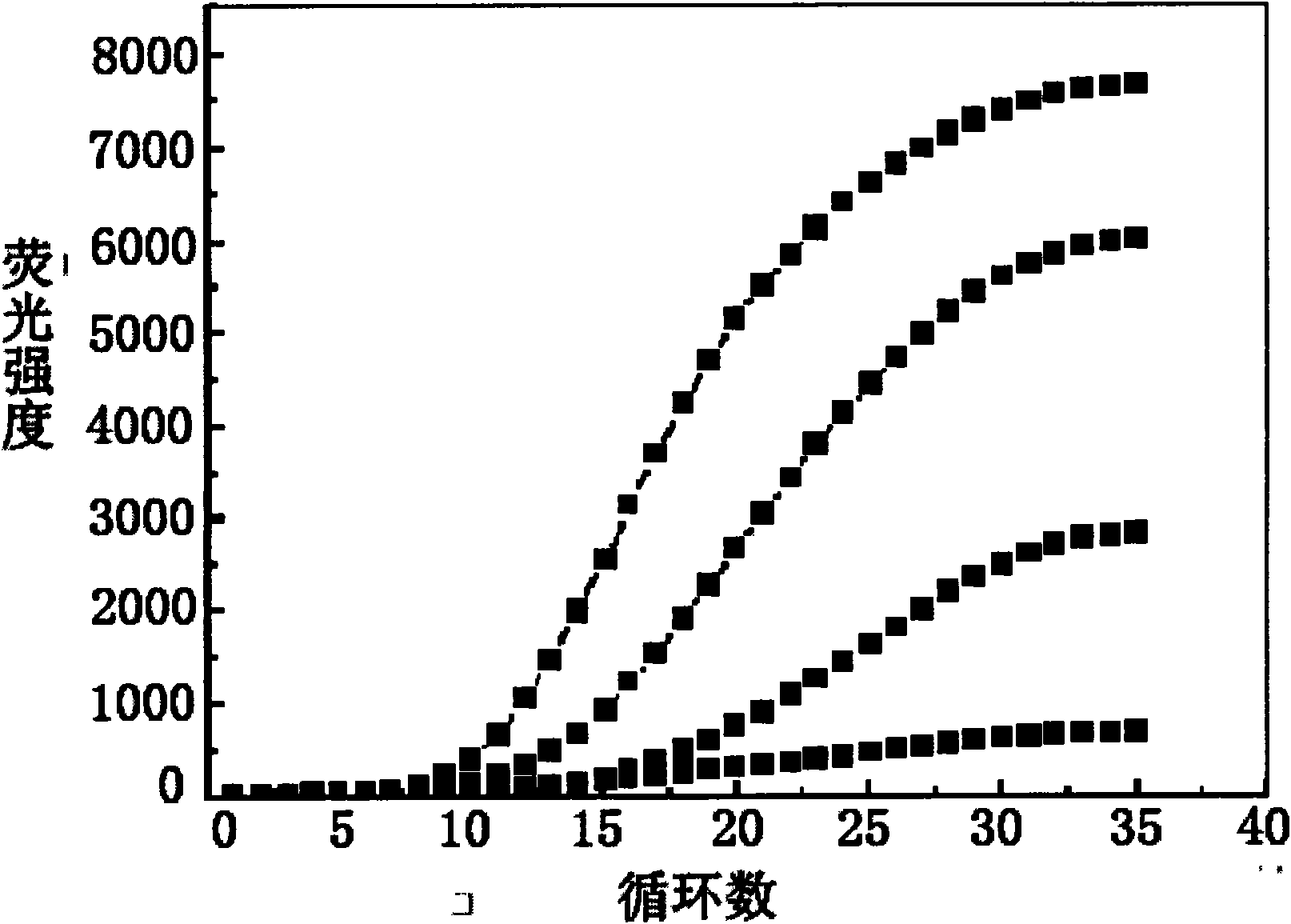
Primer design method for amplifying low-content gene mutation DNA and application thereof
A technology for primer design and primer amplification, applied in biochemical equipment and methods, DNA preparation, recombinant DNA technology, etc., can solve the problems of limited detection ability, poor selectivity, and poor specificity, and achieve increased specificity , to solve the effect of insufficient detection sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0038] In this example, a pair of primers were designed by the method of the present invention and combined with fluorescent quantitative PCR technology and double-circle probe technology to detect the gene mutation of K-ras gene 12 codon GGT>GAT.
[0039] Material:
[0040]
[0041]
[0042] 1. Preparation of samples and reference substances
[0043] 1). Sample processing and template extraction
[0044] Receive paraffin tissue samples from the clinic, cut the samples into 5-10 μm, add 1ml of xylene to dewax, collect the precipitate by centrifugation, add 1ml of absolute ethanol to the precipitate, dry at room temperature or 37°C, add proteinase K and Buffer ATL, 56 Digest and lyse at ℃ for 1 hour, incubate at 90°C for 1 hour, add 200ml Buffer AL and mix well, then add 200μl absolute ethanol and mix well, transfer the supernatant carefully to a QIA 2ml spin column, centrifuge at 6000×g (8000rpm) for 1min, add 500μl Buffer AW1, centrifuge at 6000×g (8000rpm) for 1min, ...
Embodiment 2
[0083] In this example, a pair of primers were designed by the method of the present invention in combination with fluorescent quantitative PCR technology and double-loop probe technology to detect the E746_A750del gene mutation in exon 19 of the EGFR gene.
[0084] Material:
[0085]
[0086] 1. Preparation of samples and reference substances
[0087] 1). Sample processing and template extraction
[0088] Receive paraffin tissue samples from the clinic, cut the samples into 5-10 μm, add 1ml of xylene to dewax, collect the precipitate by centrifugation, add 1ml of absolute ethanol to the precipitate, dry at room temperature or 37°C, add proteinase K and Buffer ATL, 56 Digest and lyse at ℃ for 1 hour, incubate at 90°C for 1 hour, add 200ml Buffer AL and mix well, then add 200μl absolute ethanol and mix well, transfer the supernatant carefully to a QIA 2ml spin column, centrifuge at 6000×g (8000rpm) for 1min, add 500μl Buffer AW1, centrifuge at 6000×g (8000rpm) for 1min, op...
Embodiment 3
[0128] In this example, a pair of primers were designed by the method of the present invention in combination with fluorescent quantitative PCR technology and double-circle probe technology to detect the T790M gene mutation in exon 21 of EGFR gene.
[0129] Material:
[0130]
[0131]
[0132] 1. Preparation of samples and reference substances
[0133] 1). Sample processing and template extraction
[0134]Receive paraffin tissue samples from the clinic, cut the samples into 5-10 μm, add 1ml of xylene to dewax, collect the precipitate by centrifugation, add 1ml of absolute ethanol to the precipitate, dry at room temperature or 37°C, add proteinase K and Buffer ATL, 56 Digest and lyse at ℃ for 1 hour, incubate at 90°C for 1 hour, add 200ml Buffer AL and mix well, then add 200μl absolute ethanol and mix well, transfer the supernatant carefully to a QIA 2ml spin column, centrifuge at 6000×g (8000rpm) for 1min, add 500μl Buffer AW1, centrifuge at 6000×g (8000rpm) for 1min, ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com