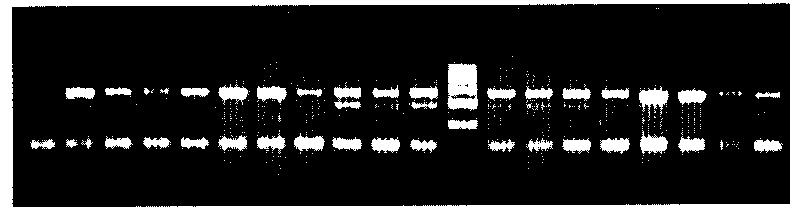
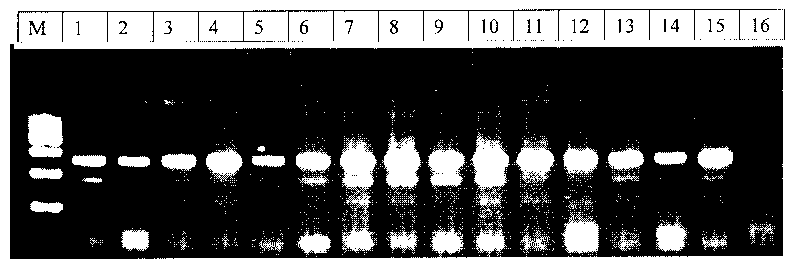
Method for rapidly detecting third exon single base mutation of myostatin gene
An exon and mutation site technology, applied in the field of molecular biology, can solve the problems of cumbersome, difficult, and time-consuming operations, and achieve the effect of simple operation, low cost, and short time
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0032] Example 1 The detection of the third exon SNP site of myostatin gene
[0033] Materials and Methods
[0034] 1. Sample collection
[0035] In this example, Piedmont cattle and Nanyang cattle crossbred offspring Pinan cattle were used as the research object for population SNPs detection. In addition, three typical representative breeds: Nanyang cattle, Piedmont cattle and Holstein cattle were selected for SNPs screening. Among them, the samples of Pinan cattle and Nanyang cattle were collected from the cattle farm of Xinye County, Nanyang District, Henan Province; the samples of Piedmont cattle and Holstein cattle were collected from the Changping Farm of the Animal Husbandry Research Institute of the Chinese Academy of Agricultural Sciences. All test samples were hair follicles, which were stored in the refrigerator after collection. The detailed test contents of each sample are shown in Table 1.
[0036] Table 1: Sources and uses of experimental animal samples
[0...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com