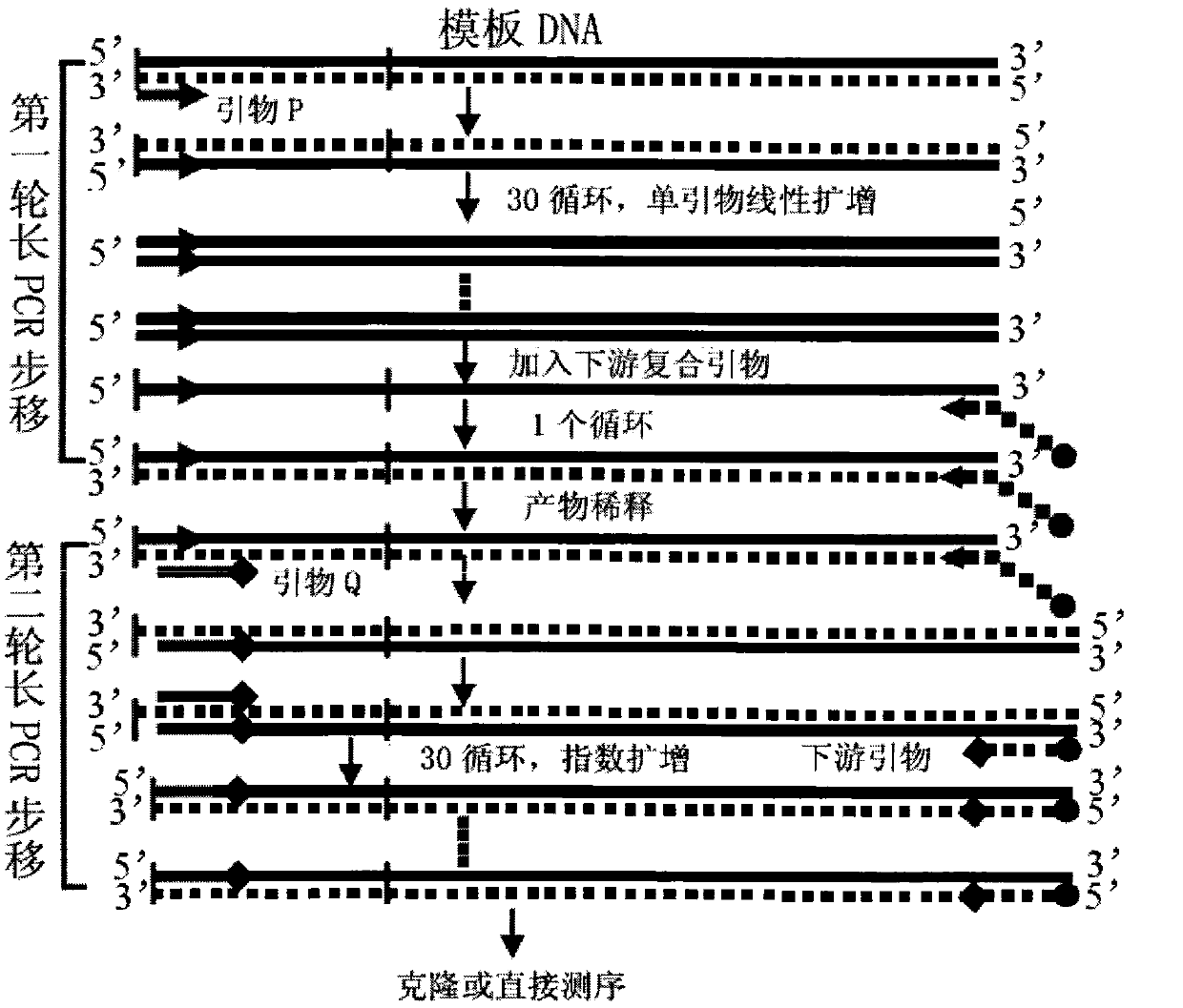
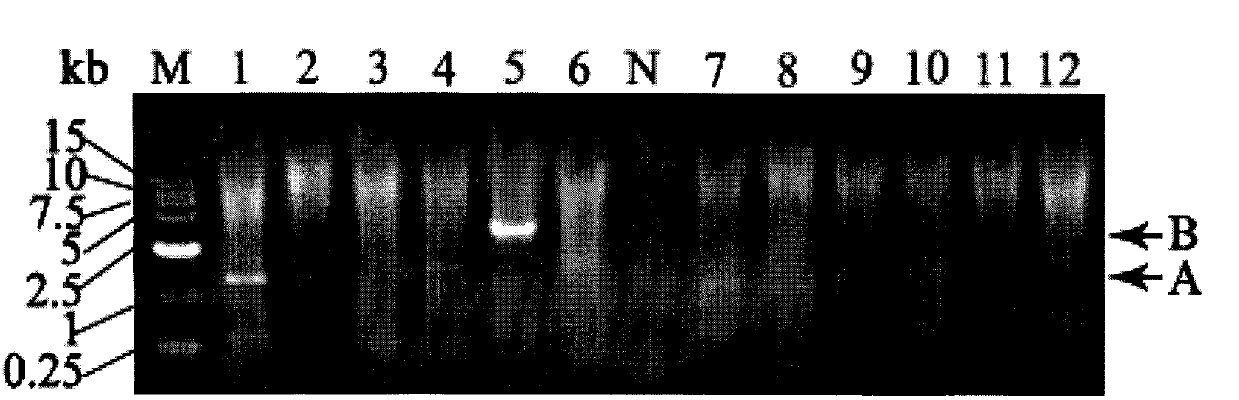
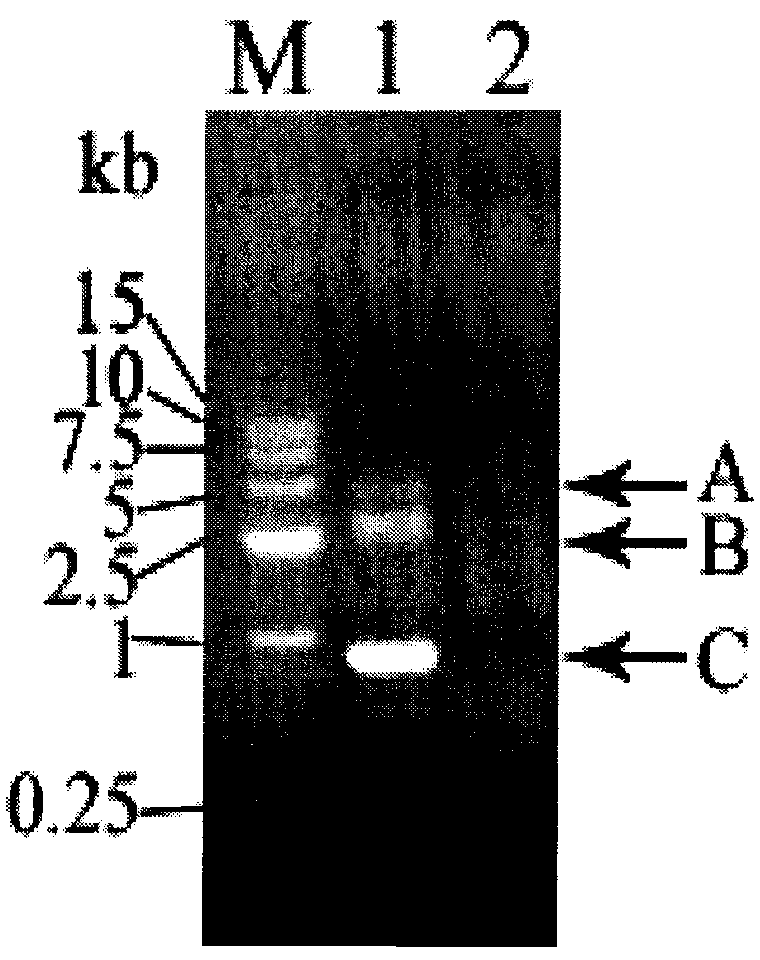
Long PCR walking kit and using method and application thereof
A kit and walking technology, which is applied in the field of long PCR walking kits, can solve the problems of cumbersome operation, complicated setting procedures, and many non-specific fragments, and achieve broad application prospects, reduced walking operations, and reduced experimental costs Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0030] Long PCR walks were used to amplify unknown DNA sequences flanking the transposon valT of Vibrio alginolyticus E0601-like Vibrio cholerae. Prepare the reagents according to the following formula.
[0031] Kit composition:
[0032] 1) Reagent A: 500 μL of the reaction premix system. Each 10.5 μL contains 1 μL dNTP (dATP, dTTP, dGTP, dCTP each 10 mmol / L), 4 μL MgCl 2 (25mmol / L), 5μL 10×PCR buffer, 0.5μL LA Taq enzyme;
[0033] 2) Reagent B: 10mL ddH 2 O;
[0034] 3) Reagent C: agarose, 6g;
[0035] 4) Reagent D: 1mL of 6×PCR product loading buffer, its components are: 30mmol / L EDTA, 36% (V / V) glycerol, 0.05% (W / V) xylene cyanine, 0.05% (W / V )BPB.
[0036] 5) Reagent E: Goldview nucleic acid dye, 40 μL;
[0037] 6) Reagent F: 10 mL of 50×TAE buffer solution, containing 242 g of Tris, 37.2 g of ETDA, and 57.1 mL of acetic acid per 1 L.
[0038] 7) Primer series, concentration 10 μmol / L, 10 μL each. The sequence is shown as SEQ ID NO: 1-10;
[0039] 8) One primer,...
Embodiment 2
[0052] According to the partial sequence of the rpoB gene of Vibrio vulnificus 1.1758, the nearly full-length gene of rpoB was amplified by long PCR walking. Prepare reagents according to the following formula:
[0053] 1) Reagent A: 300 μL of reaction premix system. Each 11.4 μL reagent A contains 2 μL dNTP (dATP, dTTP, dGTP, dCTP each 10 mmol / L), 4 μL MgCl 2 (25mmol / L), 5μL 10×PCR buffer, 0.4μL LATaq enzyme;
[0054] 2) Reagent B: 20mL ddH 2 O;
[0055] 3) Reagent C: agarose, 10g;
[0056] 4) Reagent D: 1 mL of 6×PCR product loading buffer, its components are: 25mmol / L EDTA, 30% (V / V) glycerol, 0.06% (W / V) xylene cyanide, 0.04% (W / V )BPB;
[0057] 5) Reagent E: Goldview nucleic acid dye, 30 μL;
[0058] 6) Reagent F: 30 mL of 50×TAE buffer solution, containing 242 g of Tris, 37.2 g of ETDA, and 57.1 mL of acetic acid per 1 L;
[0059] 7) Primer series, concentration 15 μmol / L, 10 μL each. The sequence is shown as SEQ ID NO: 1-10;
[0060] 8) One primer, 15 μL, conc...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com