A pair of cabbage turnip mosaic virus EST-SSR markers and application thereof
A technology of turnip mosaic virus and Chinese cabbage, which is applied in the direction of DNA/RNA fragments, measurement/inspection of microorganisms, recombinant DNA technology, etc., can solve problems such as difficult to distinguish homozygous and heterozygous, achieve high accuracy and speed up The effect of selective breeding and efficient selection
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0027] (1) Using the improved CTAB method to extract the genomic DNA of the single plant to be tested
[0028]In this example, the Chinese cabbage inbred line material "73" with high resistance to TuMV and the inbred line material "71-36-2" with high TuMV susceptibility were selected as parent materials, and the national-level resistant source of TuMV "8407" was selected. The core germplasm material "Guan 291" sensitive to TuMV was used as the control material. Take 0.1 g of the young leaves of the above materials, grind them into powder in liquid nitrogen, and put them into a 1.5 mL centrifuge tube. Add 700 μL of 65°C preheated CTAB extraction buffer (2.0% CTAB (g / ml); 1.4mol / L NaCl; 1mol / L Tris-HCl, pH=8.0; 0.5mol / L EDTA; 1.0% β-mercaptoethanol (V / V)), mix well, bathe in water at 65°C for 45min-1h, and shake once every 5-10min. Cool to room temperature, centrifuge to take the supernatant, extract with an equal volume of Tris-saturated phenol:chloroform:isoamyl alcohol (25:...
Embodiment 2
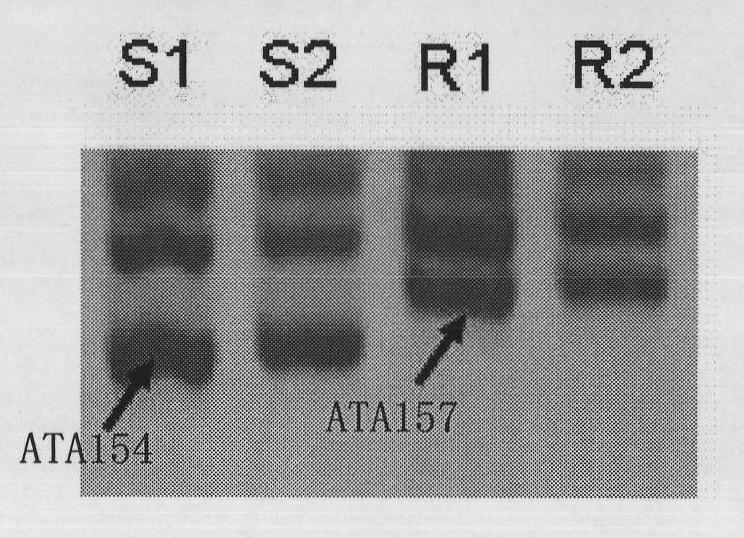
[0035] Such as figure 1 As shown, using the primer HCC259, a 157bp band was amplified in the resistant parental material "73" and the disease-resistant control material "8407", named ATA157, and in the susceptible parental material "71-36-2" A 154bp band was amplified in the control material "Crown 291" and the susceptible control material, named ATA154.
Embodiment 3
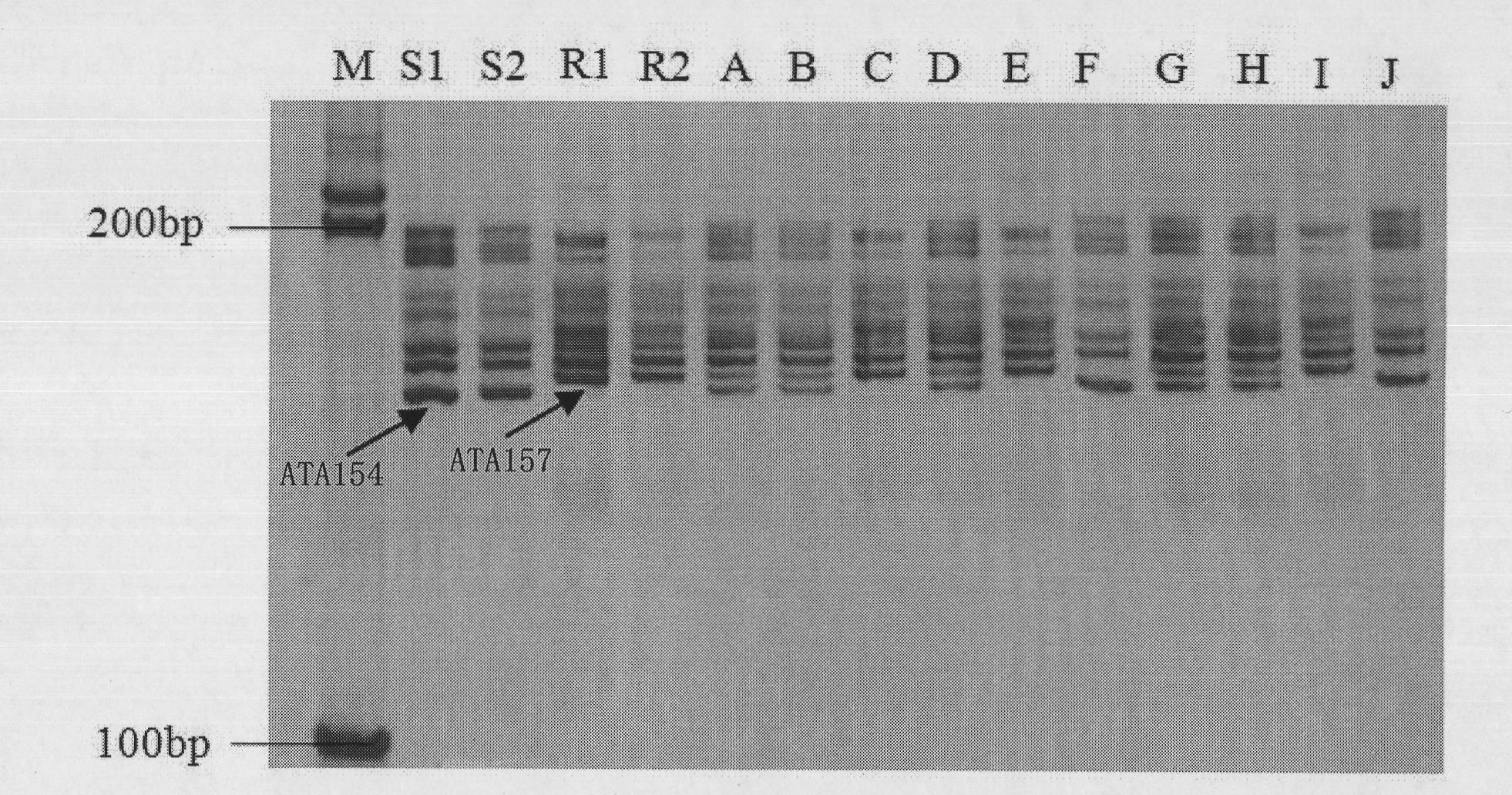
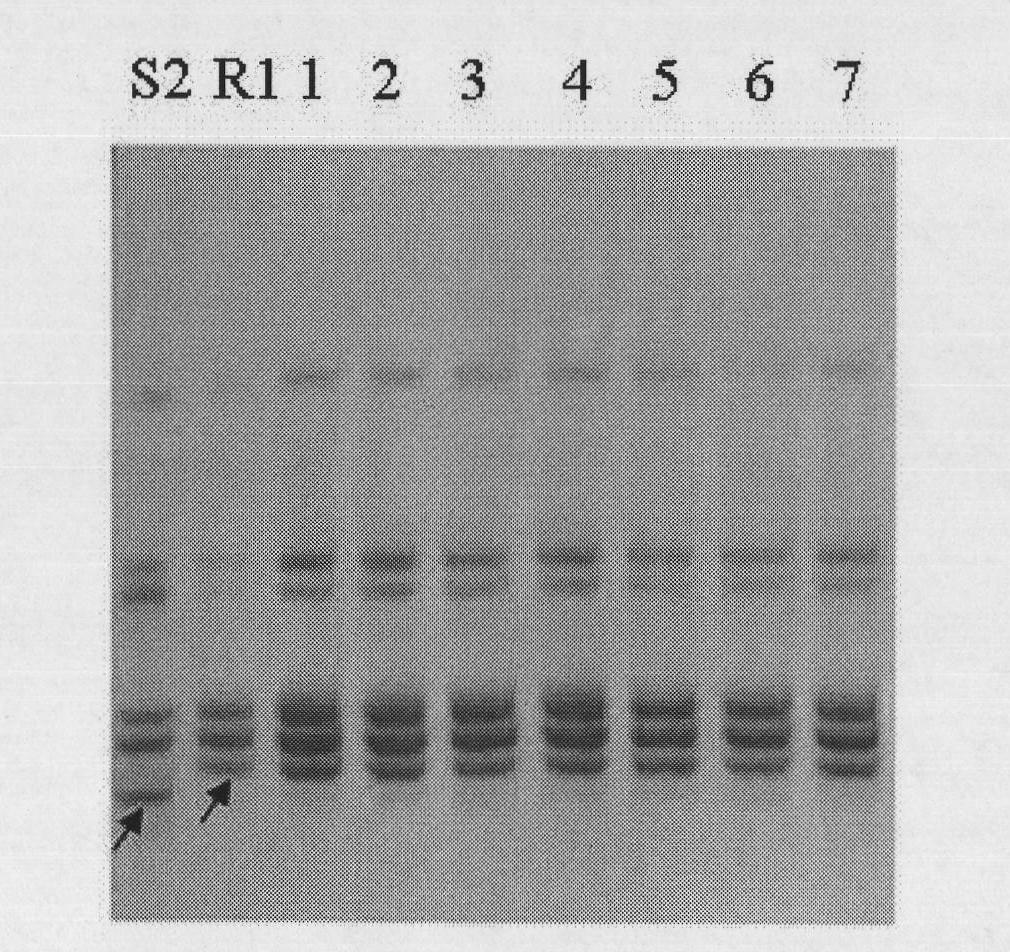
[0037] Such as figure 2 As shown, using primer HCC259 to detect 10 individual plants (A-J) of F2 generation isolated populations, wherein, some individual plants (such as No. C, E, and No. , J individual plants) the susceptible band ATA154 was expanded, and some (A, B, D, G, H individual plants) expanded the heterozygous band.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com