Cellulose degrading enzyme with glucosidase/xylosidase dual functions and preparation method and application thereof
A technology of cellulose degrading enzyme and glucosidase, which is applied in the field of bioengineering to achieve the effects of reducing viscosity, high product purity and increasing digestibility
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0052] Example 1 Extraction of Chinese Yak Rumen Microbial Metagenomic DNA
[0053] Collect rumen samples from yaks in Qinghai, China, filter them with 3 layers of gauze, and centrifuge the filtrate to collect rumen microbial cells, take 100-200ul bacterial samples, wash 2-3 times with PBS, add 650uL DNA extraction buffer (Tris-HCl, 100mM pH8 .0; Na 2 EDTA 100mM pH8.0; Na 3 PO 4 Buffer 100mMpH8.0; NaCl 1.5M; CTAB 1%; pH8.0), mix well, place at -80°C, then place in a 65°C water bath to melt, repeat three times; add 3-4μL lysozyme (100mg / L) Shake horizontally (37°C, 225rpm) in a shaker for about 30min; add 2-3μL proteinase K (20mg / mL) and continue shaking for about 30min; add 50-70uL SDS (20%), after mixing, 65 Incubate at ℃ for 1-2 hours, mix the centrifuge tube up and down every 10-20 minutes; centrifuge at 12,000 rpm for 10 minutes at room temperature, collect the supernatant, add 400-500ul of phenol: chloroform: isoamyl alcohol (25:24:1) for extraction Twice; extract o...
Embodiment 2
[0054] Example 2 Construction of yak rumen microbial metagenomic cosmid library and screening of glucosidase activity
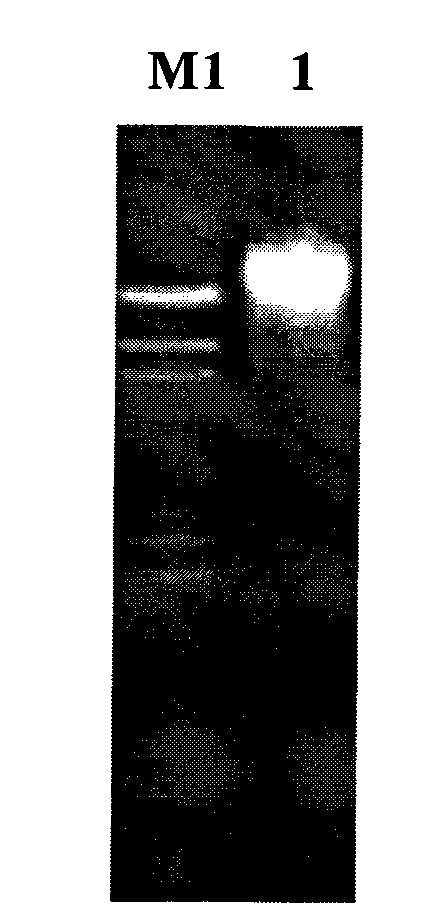
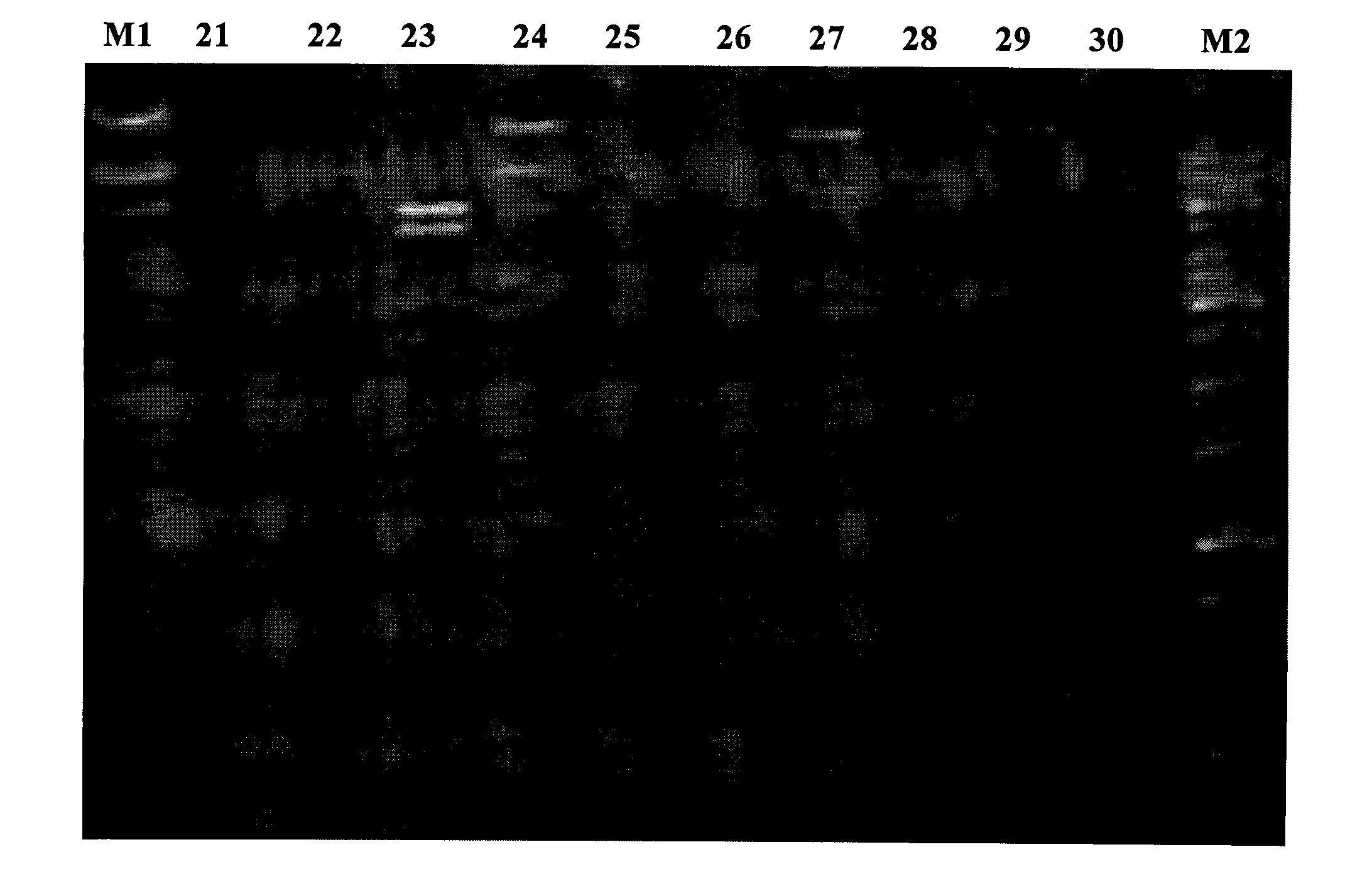
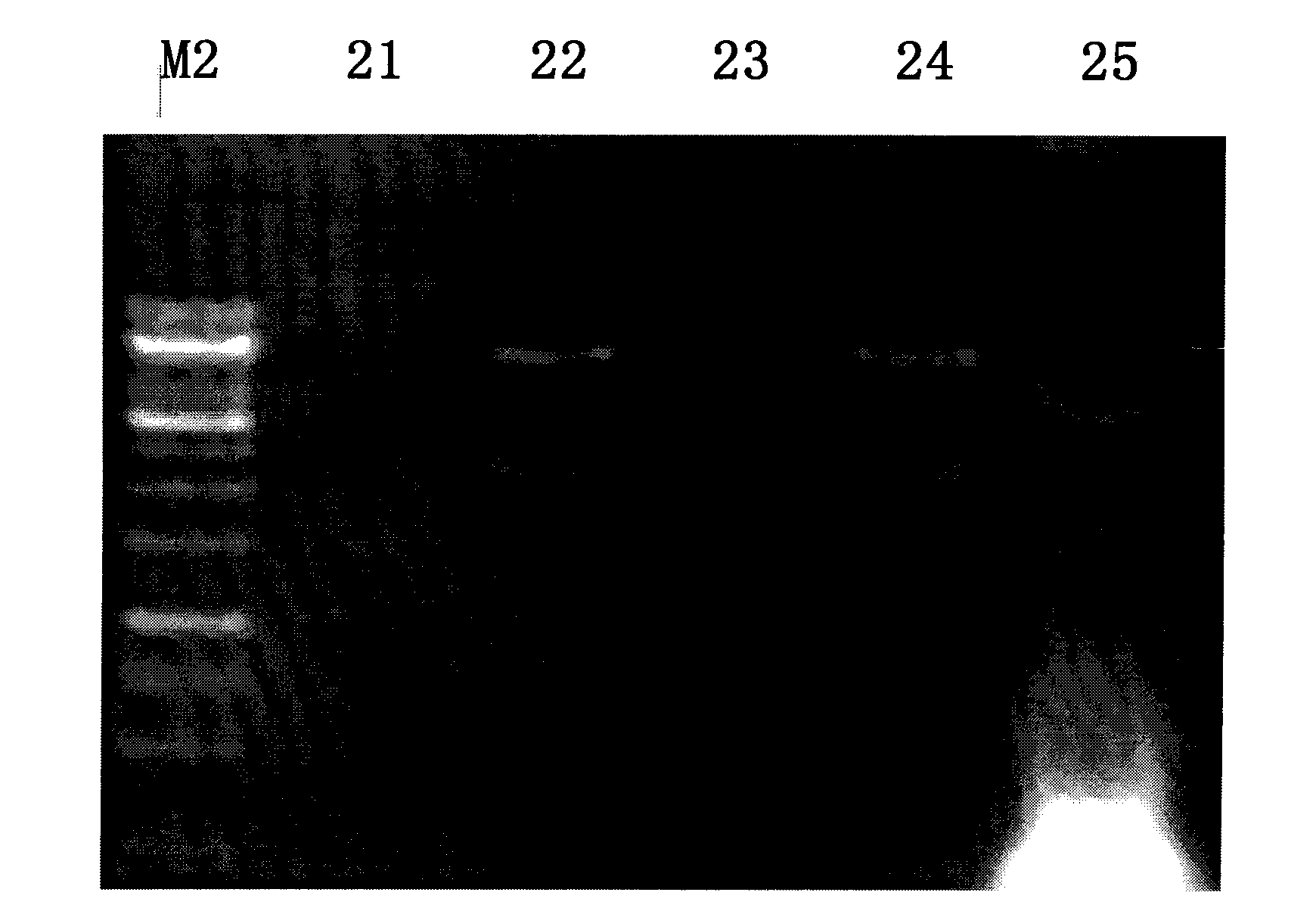
[0055] For the construction method of the rumen-uncultured bacterial metagenomic library, refer to the product manual of the pWEB::TNC Cosmid Cloning Kit kit from Epicentre Company. The extracted metagenomic DNA was blunted with End-Repair Enzyme Mix, ligated with the dephosphorylated vector pWEB::TNC in the kit, and the ligated product was used MaxPlax TMAfter the Lambda Packaging Extracts were packaged, the host bacteria E.coli EPI100 was infected, and the infected products were spread on the plate, and colonies were grown after being incubated at 37°C for 12-16 hours, which was the metagenomic library of this batch of samples. Randomly call 10 cosmid clones to extract plasmid DNA, digest and analyze with BamH I (such as figure 2 ), the results showed that the quality of the constructed library was good, and the size of the inserted genome fragment was ab...
Embodiment 3
[0057] Cloning and analysis of the sequence of the RuBGX1 gene of embodiment 3 rumen microorganism source
[0058] The screened cosmid-positive library clones were subcloned in pGEM11z, functionally screened, and specific oligonucleotides were used as sequencing reaction primers to determine the RuBGX1 gene from rumen microorganisms, its promoter / regulator, and the structure of the gene Sequences of part and terminator regions.
[0059] In order to determine the nucleotide sequence, the isolated part of the Sau3A I fragment was fragmented (2-3kb), and then it was cloned into the vector pGEM11z, and 4-methylumbelliferyl β-D-1,4-glucoside was used as The substrates were screened for the function of β-glucosidase in the sub-library, and the nucleotide sequences of the obtained positive sub-library clones were determined by T7 sequencing primers and SP6 sequencing primers of the vector pGEM11z. The coding region sequence analysis was performed using the analysis software DNAMAN. ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com