Application of three microRNAs in diagnosis and treatment of human acute myeloid leukemia
A technology for acute myeloid and leukemia, applied in gene therapy, microbial determination/examination, biochemical equipment and methods, etc., can solve problems such as no research report yet
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0037] Embodiment 1: Experimental object
[0038] All AML patients were selected from the visiting patients by the Department of Hematology, Affiliated Hospital of Inner Mongolia Medical College. The diagnosis of AML was based on conventional methods, and the morphological diagnosis and typing of each case were in line with the requirements of the "Standards for the Diagnosis and Curative Effect of Hematological Diseases". The age and gender-matched normal controls were selected from volunteer blood donors. In addition to normal hemogram indicators to exclude blood diseases, other important diseases were also excluded. All followed the principle of informed consent. The research plan was approved by the ethics review committees of both parties (Institute of Basic Medical Sciences, Chinese Academy of Medical Sciences and the Affiliated Hospital of Inner Mongolia Medical University).
Embodiment 2
[0039] Example 2: Detection of Abnormally Expressed miRNAs in AML Patients Using miRNA Chip Analysis
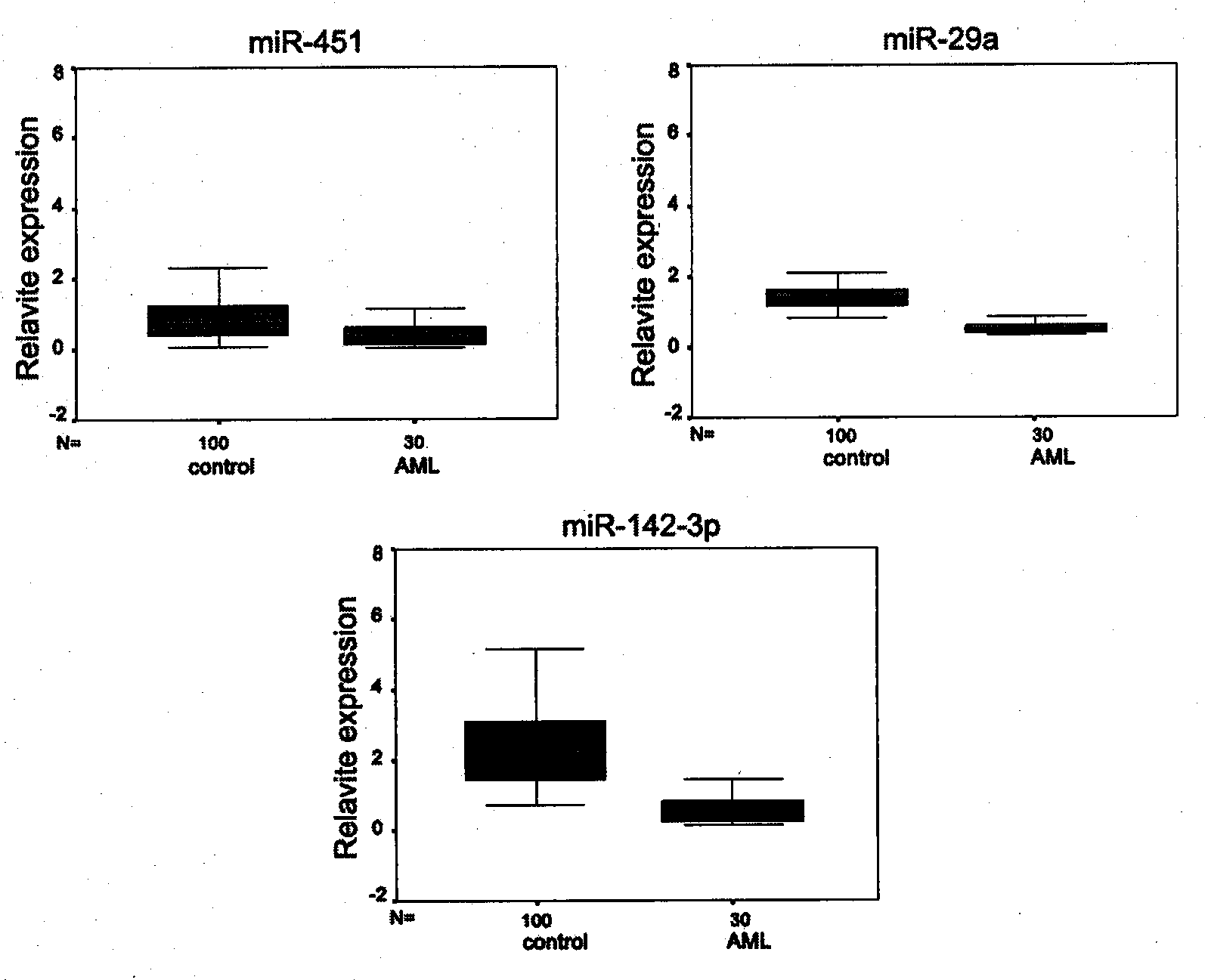
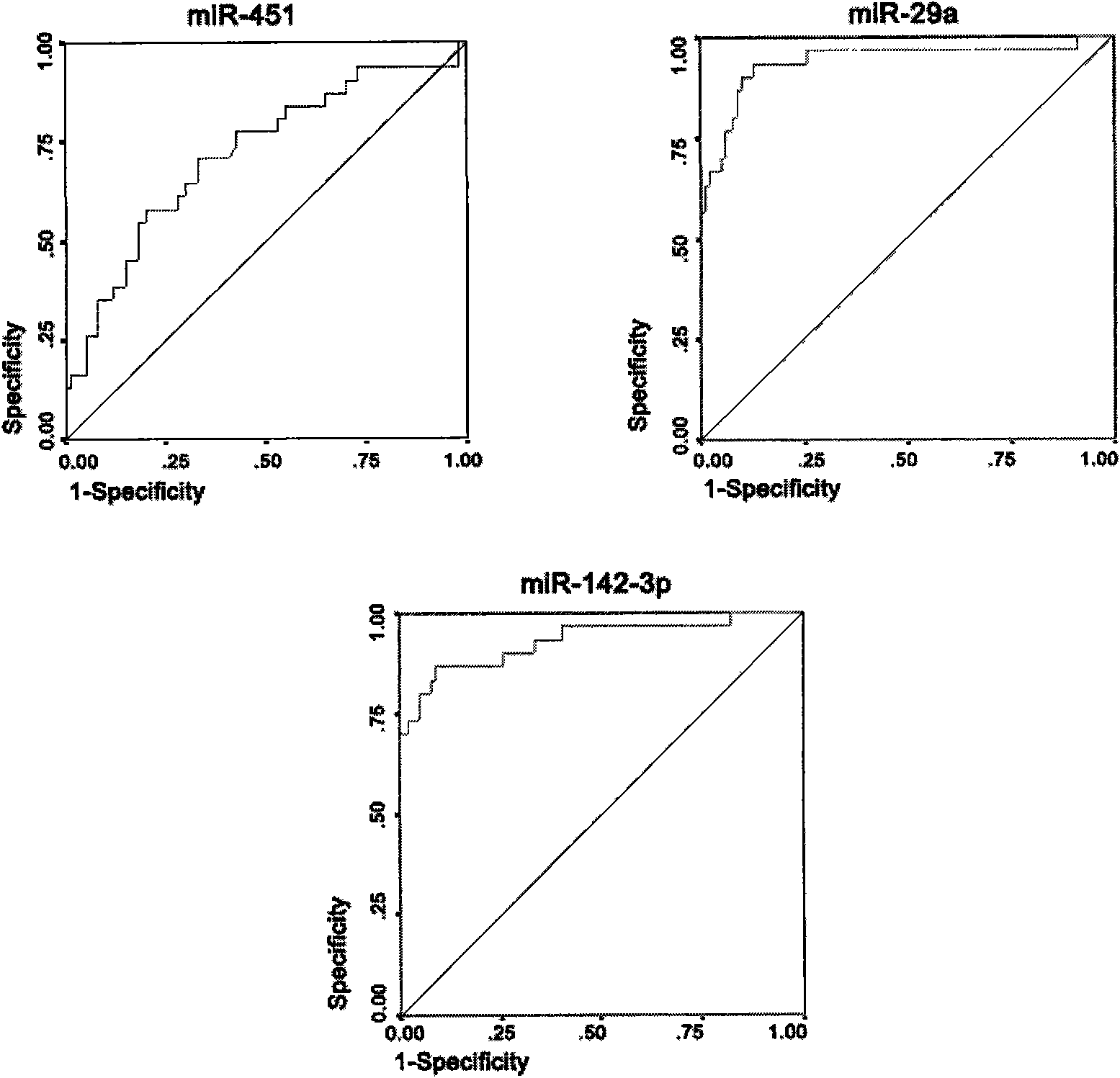
[0040] In order to compare the expression differences of miRNAs in AML patients and normal controls, we first used the miRNA microarray analysis method. The samples used for microarray analysis were 2 cases of newly diagnosed AML (M5 type) cases and 2 cases of normal controls matched in gender and age. Isolate the mononuclear cells of its peripheral blood (the separation method is described in detail in the next paragraph), and entrust Shanghai Kangcheng Bioengineering Co., Ltd. to perform the miRNA chip experiment operation (using Exiqon, LNA TM microRNA chip) and provide complete experimental and analysis reports.
[0041] Peripheral blood mononuclear cells (MNC) were isolated by density gradient centrifugation. The collected fresh peripheral blood was diluted with 1×phosphate-buffered saline (PBS) containing 2mmol / L EDTA according to the volume of 1:4, and the pipette wa...
Embodiment 3
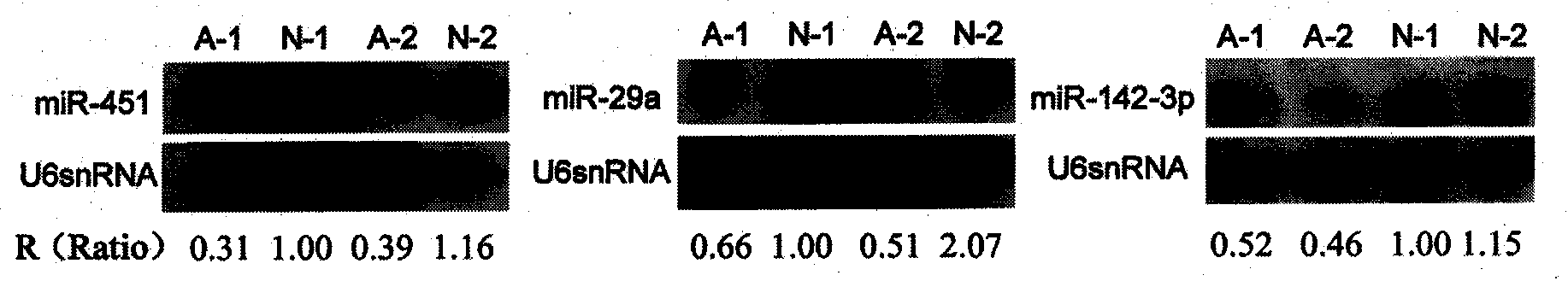
[0042] Example 3: Northern hybridization verification of chip results
[0043] In order to eliminate the false positive results in chip analysis, we verified some of the results by Northern hybridization. Total RNA is first extracted from a mononuclear cell sample. at approximately every 1 x 10 7 1 ml Trizol (purchased from Invitrogen) was added to each mononuclear cell, and the cells were immediately repeatedly pipetted and left at room temperature for 5 min to lyse. Add 0.2 ml of chloroform to the lysis pipette, vibrate the centrifuge tube on a vortex shaker for 15 s, let it stand at room temperature for 2-3 min, and then centrifuge at 12,000×g for 15 min at 4°C. Transfer the upper aqueous phase to a new centrifuge tube, add 0.5ml of isopropanol to mix, and place at room temperature for 10 minutes. Centrifuge at 12,000×g for 10 min at 4°C and discard the supernatant. Rinse the precipitate once with 75% ethanol, centrifuge at 7500×g for 5 min, discard the supernatant, and...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Density | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com