Traceless modification method of chromosome
A modification method and chromosome technology, applied in the field of chromosome genome modification, can solve the problems of no positive screening, time-consuming labor intensity, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
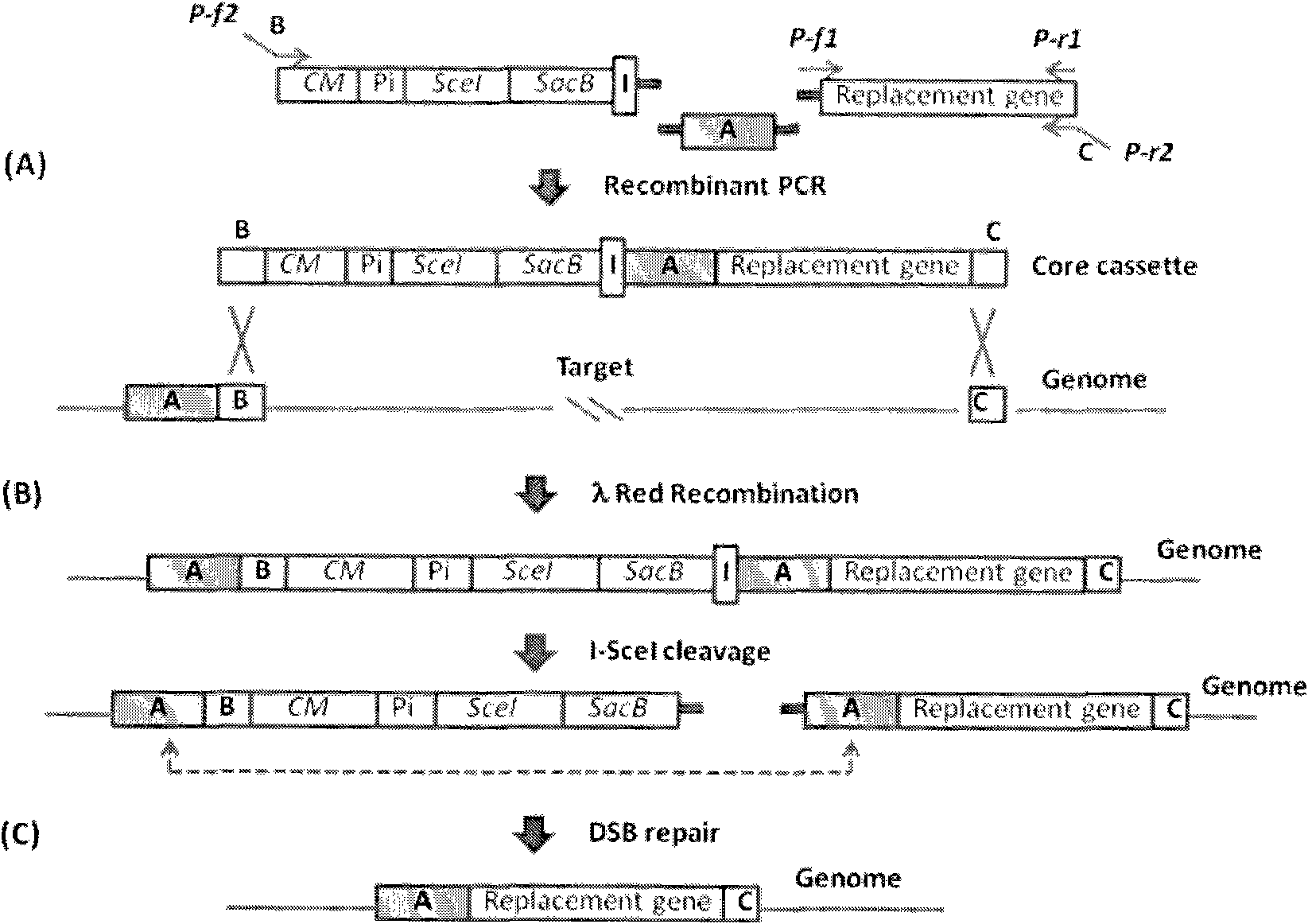
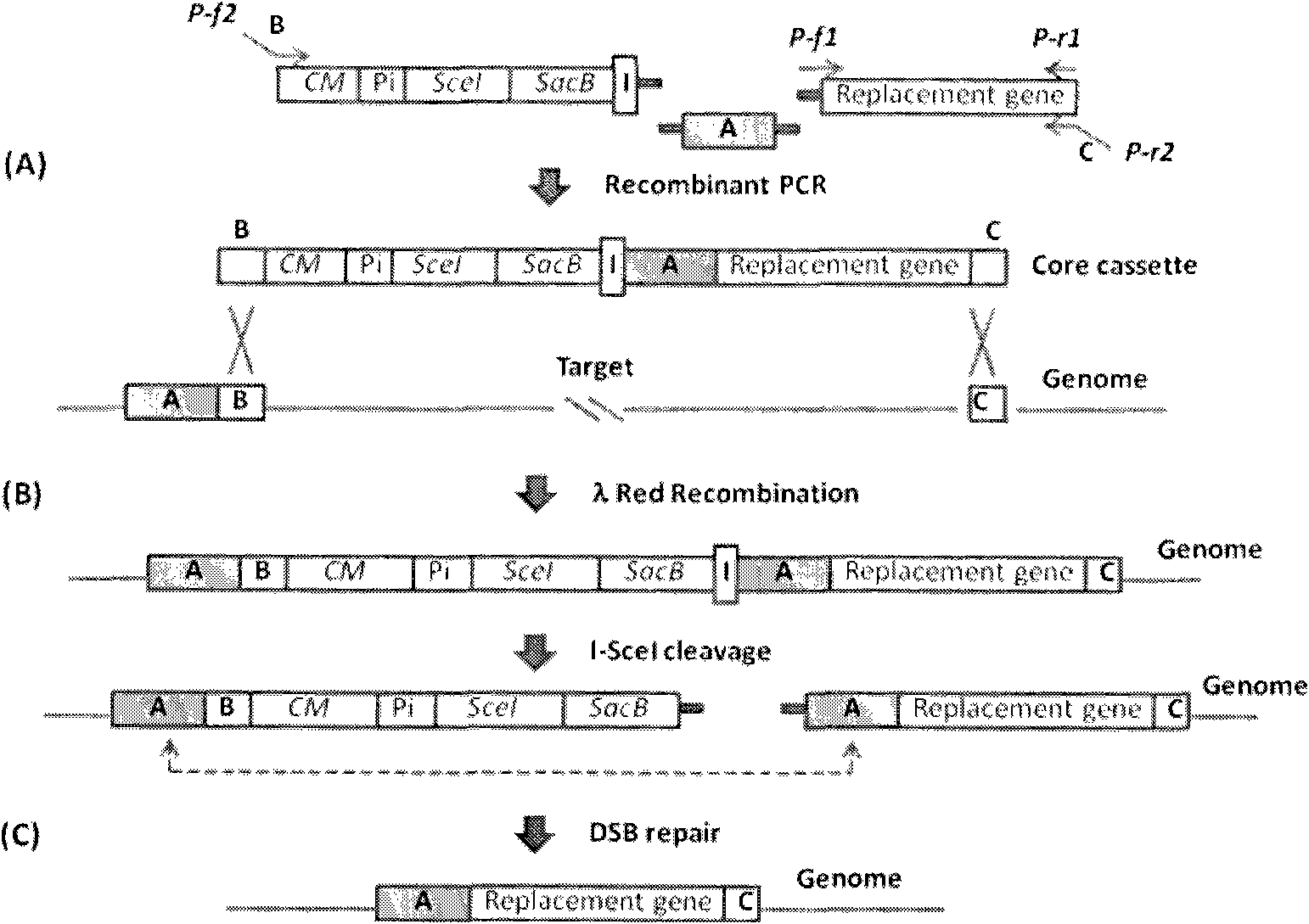
[0024] The present invention seamlessly modifies host chromosomes by using positive selection and reverse selection markers. First, the target gene is replaced with a DNA fragment containing a positive resistance marker, recombinants are detected in selective medium, and then selected by I-SceI-induced double-strand breaks and counter-selectable marker gene sacB without any selectable marker or foreign DNA The final recombinant of the sequence.
[0025] According to the DNA recombination and selection mechanism in the target cell body, the present invention reduces some unnecessary steps such as plasmid transformation and repeated screening.
[0026] The present invention uses a counter-selectable marker gene, such as the sacB gene of Bacillus subtilis, to make a large number of bacteria sensitive to sucrose. The SacB gene encodes fructan sucrase, which catalyzes the hydrolysis or polymerization of sucrose to form the lethal product fructan. E. coli and many Gram-negative ba...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com