Method for designing microRNA probe sequence
A probe sequence and design method technology, applied in the field of microRNA probe analysis, can solve the problems of inability to use microRNA probe sequence design, limiting the value of microRNA chips, and low accuracy.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
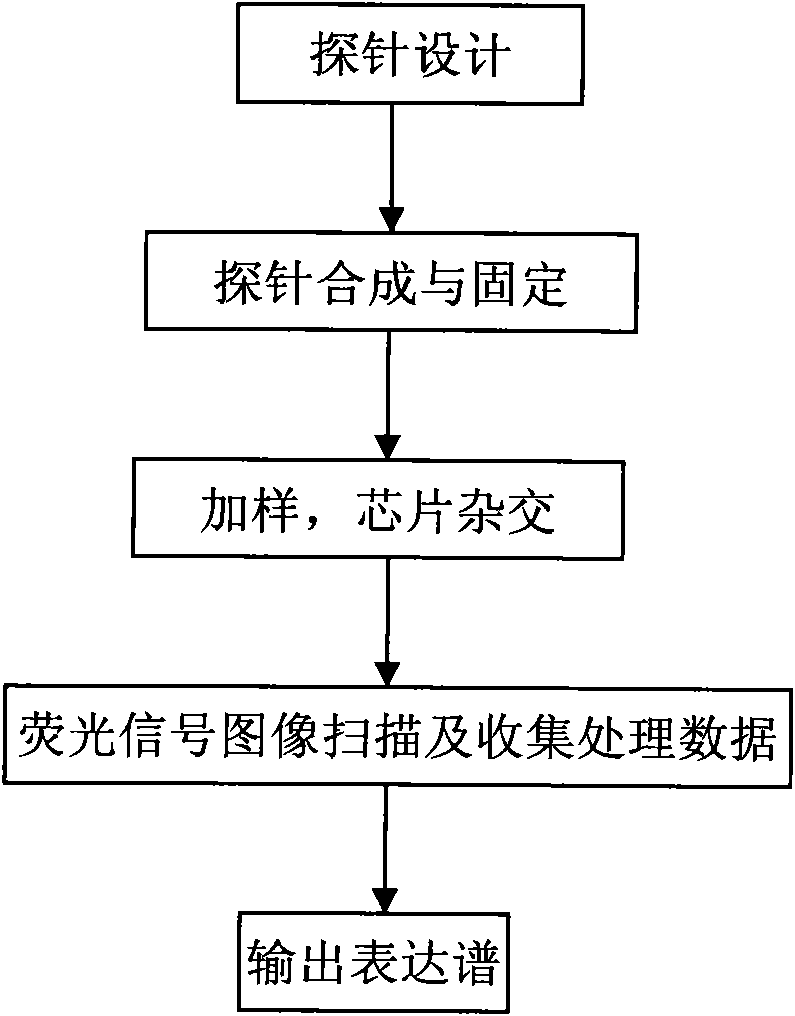
[0040] Such as figure 1 As shown, the chip probes are designed according to the detection requirements according to the database and the calculation method to analyze the microRNA mature body sequence, and the internal standard and external standard probes are designed; the probes are synthesized on the substrate or the probes are synthesized and then spotted with a spotter Fixed on the substrate of the chip; after fluorescently labeling the sample to be tested, the sample is added to the high-throughput biochip detection device, hybridized with the microRNA probe on the chip, and cleaned to remove impurities after hybridization; photoelectric detection, such as Fluorescence signal image scanning, recording microRNA expression intensity data, and preprocessing the data to generate microRNA expression profile data.
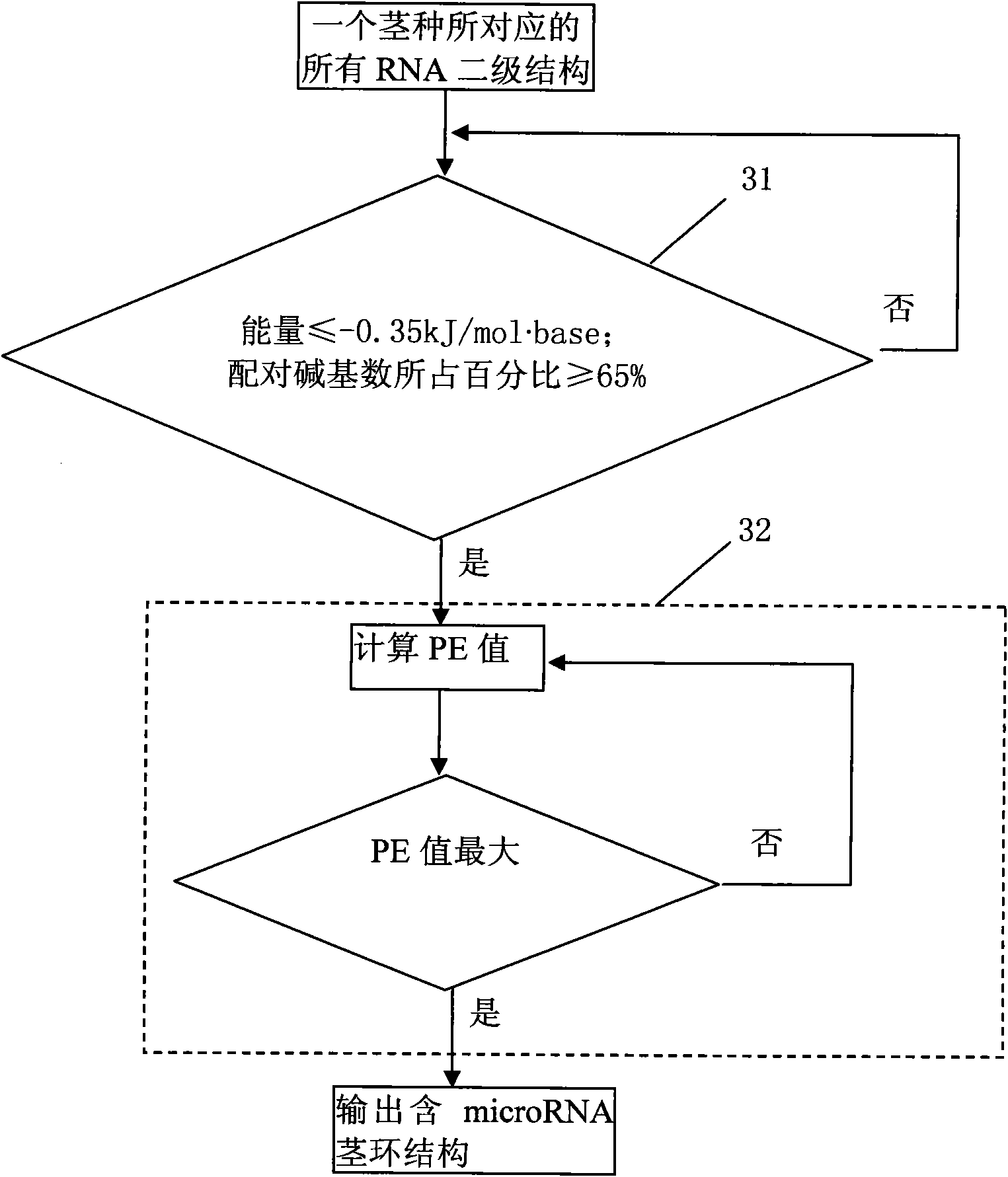
[0041] The method of the embodiment of the present invention is specifically as follows figure 2 Shown:
[0042] Step 1. Input the nucleic acid sequence of the genome,...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com