Genome-wide construction of schizosaccharomyces pombe heterozygous deletion mutants containing gene-specific barcodes by the methods of 4-round serial or block pcr, or total gene synthesis thereof
A fission yeast, barcode technology, applied in genetic engineering, plant genetic improvement, botanical equipment and methods, etc., can solve the problems of low success rate, low homologous recombination rate of fission yeast, difficulty in strain preparation, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
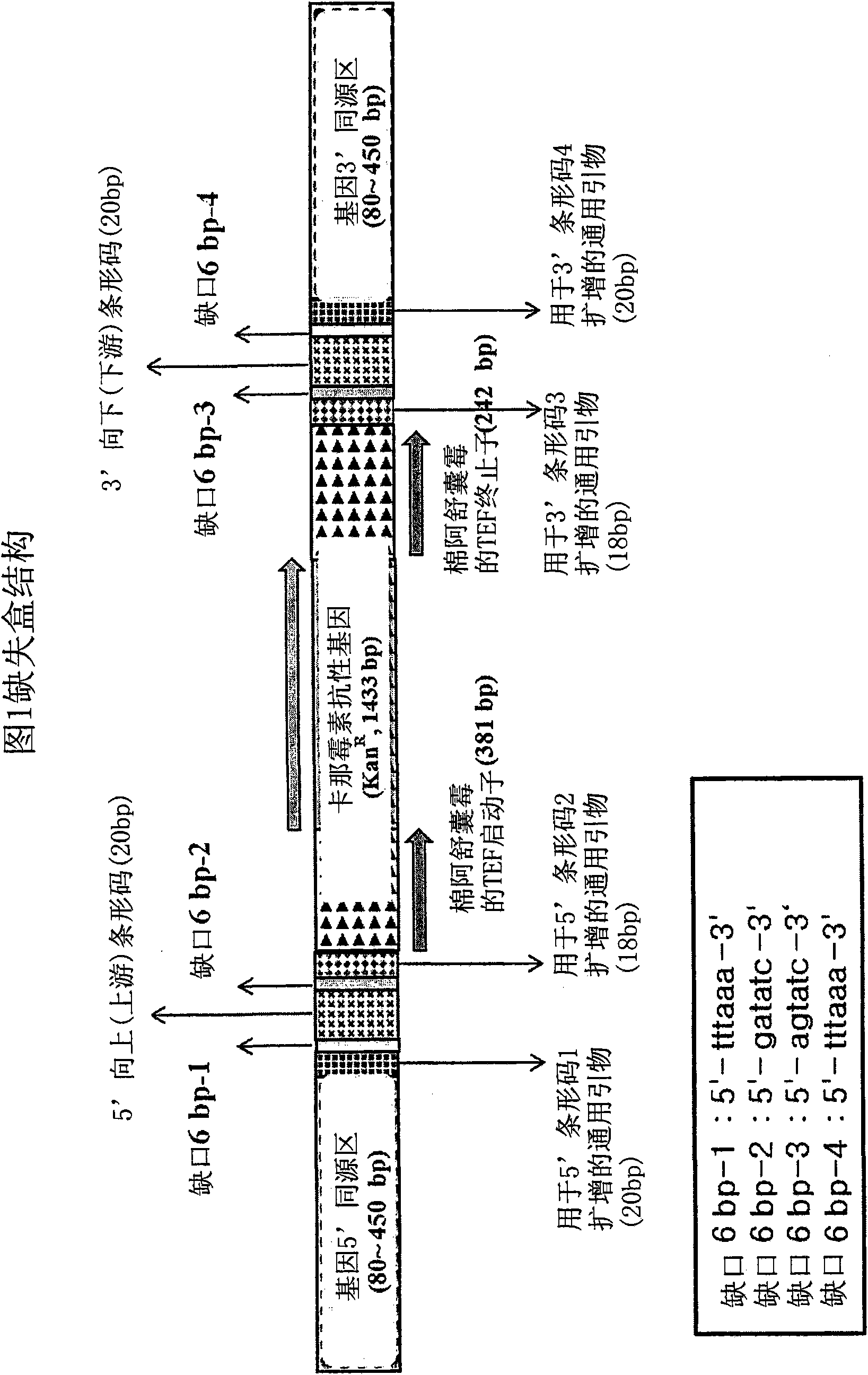
[0092] Example 1: Construction of deletion cassettes by four rounds of serial PCR or blocking PCR
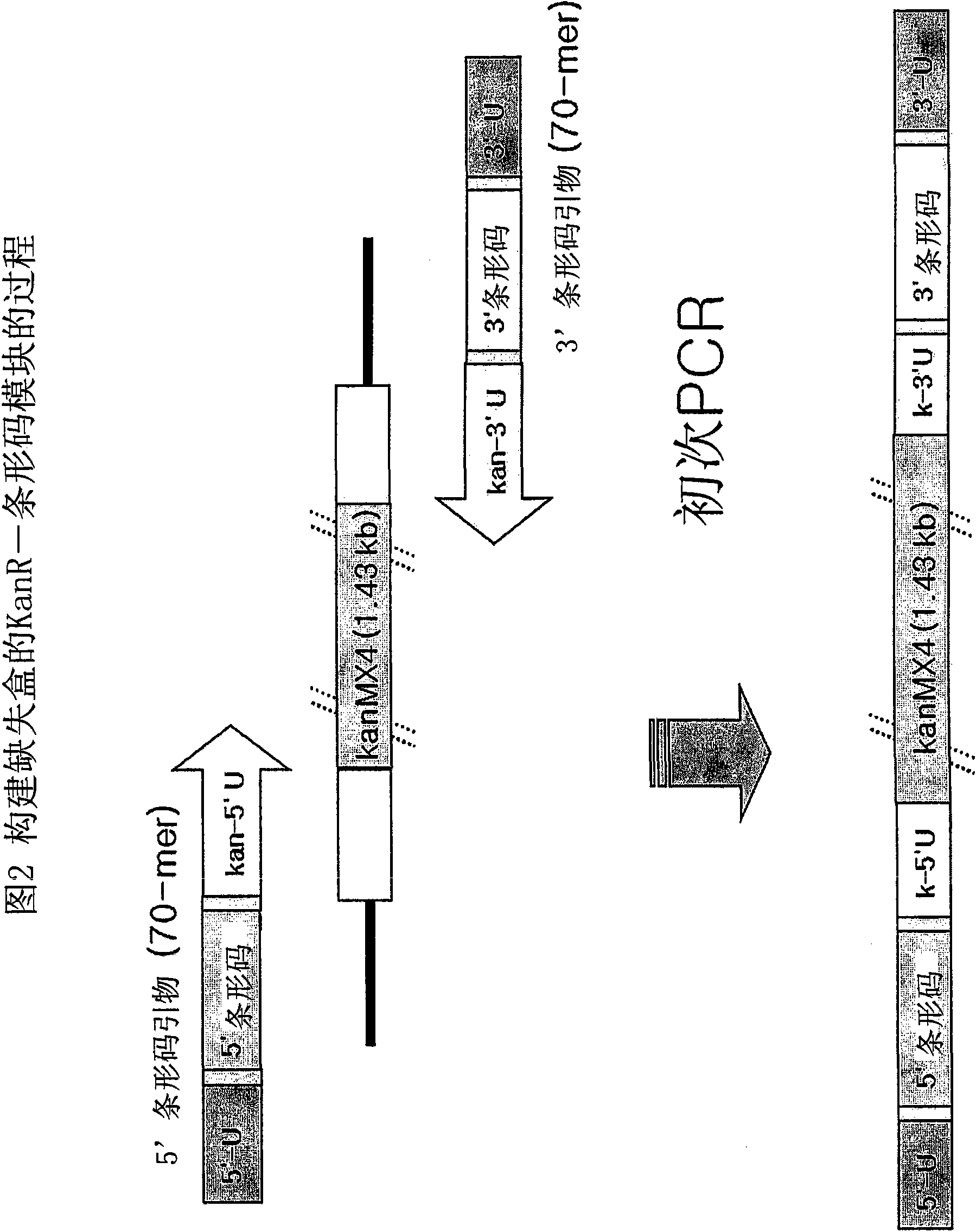
[0093] Deletion cassettes can be constructed in two different PCR techniques: four rounds of serial PCR and blocking PCR. The two PCR-based methods were pre-prepared Kan r Common among barcode modules. The base sequence of KanMX4 is well known in the art and will not be specifically described. Primary PCR was performed with a pair of 70-mer 5′ and 3′ barcode primers, each containing a gene-specific barcode, and KanMX4 served as a template to prepare Kan r Barcode module ( figure 2 ). After electrophoresis on an agarose gel stained with ethidium bromide (EtBR), the Kan thus prepared was extracted from the gel by excision under UV light. r Barcode module.
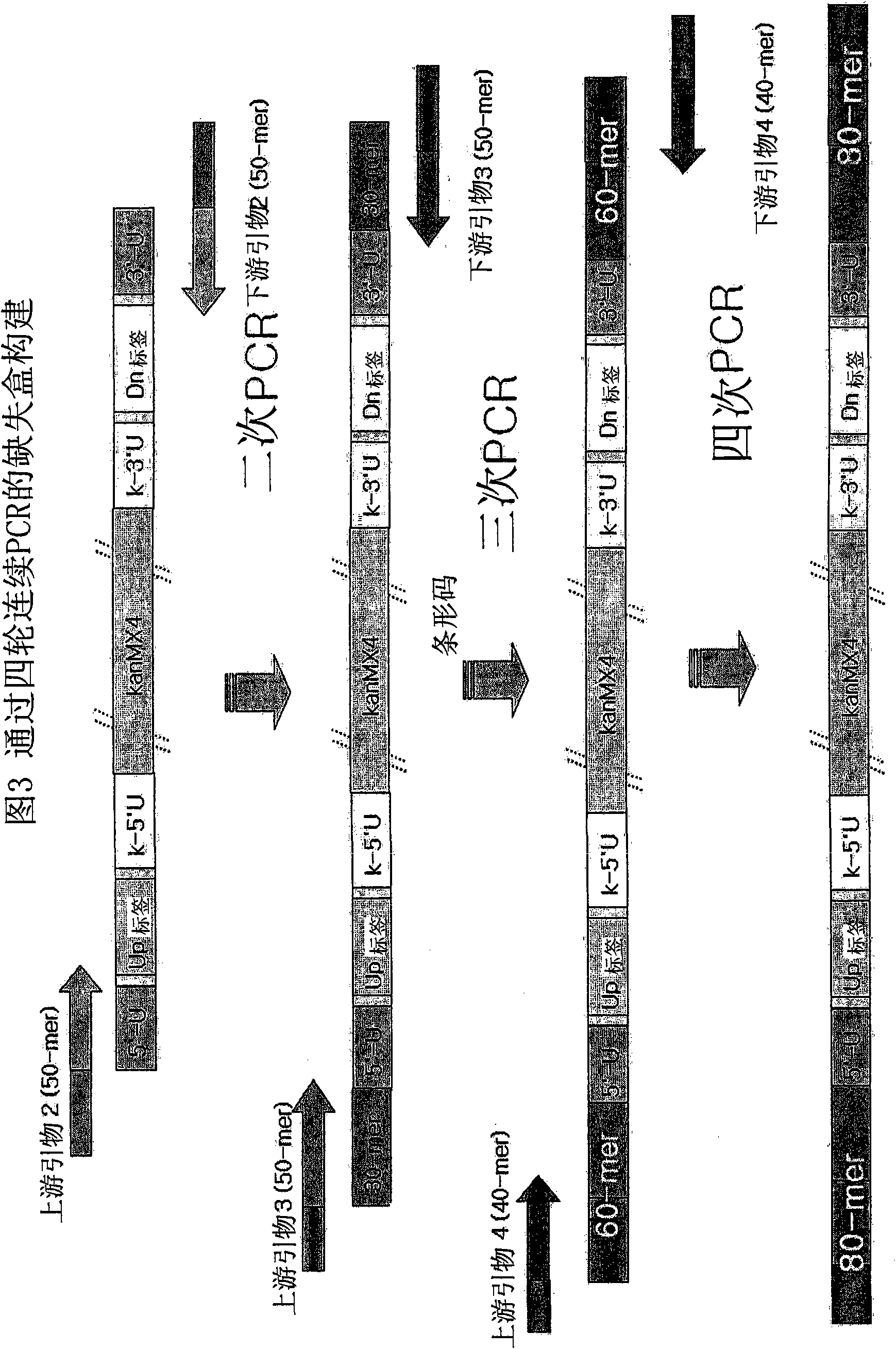
[0094] The four rounds of serial PCR used to construct the deletion cassette were similar to the conventional PCR used to construct gene-targeted budding yeast or fission yeast mutants, but differed by the addition of ...
Embodiment 2
[0096] Example 2. Construction of deletion cassettes by gene synthesis
[0097] Even the PCR technique described above failed to generate gene-targeted mutants up to 4% of all genes. The reason is because the high content of adenine (A) and thymine (T) in the promoter and terminator of the gene does not allow primer sites for conventional PCR. Taking this issue into consideration, a gene synthesis method was utilized. The present invention is the first to use gene synthesis in constructing deletion cassettes and making deletion fission yeast mutants.
[0098] The deletion cassette was designed to consist of three fragments: 1) 5' chromosome homology region and 5' barcode with 5' common linker sequence (common likersequence), 2) kanamycin resistance gene (Kan 1 ), and 3) 3' barcodes and 3' chromosomal homology regions with 3' common linker sequences. To join the three fragments into a row, overlapping junction oligonucleotide sequences were arranged between the fragments t...
Embodiment 3
[0101] Example 3: Transformation of deletion cassettes and identification of gene-targeted mutants
[0102] Using the lithium acetate method, the deletion cassettes prepared by Examples 1 and 2 were transformed into diploid S. ) (Moreno et al., Methods Enzymol. 194:795-823). Spread the transformed strains on YES agar plates and culture them at 30°C for 3-4 days to form colonies. Colony PCR was performed to check if the desired gene was targeted. Pick a small number of cells from the edge of the growing colony on the agar plate and suspend them in deionized water. A portion of the suspension was subjected to PCR with a pair of primers. Such as Figure 6 Colony PCR was performed using a pair of CP5 and CPN1 or CPN10 for the 5' side of the deletion cassette for insertion into the chromosome, and a pair of CP3 and CPC1 or CPC3 for the 3' side of the deletion cassette as indicated. CP5 and CP3 are located at 100-200 bp upstream and downstream of the homologous regions of the...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com