Method for detecting piwi-interacting ribonucleic acid (piRNA) in gastric juice
A detection method and gastric juice technology, applied in the field of piRNA detection in gastric juice, can solve the problems of low piRNA, no specific extraction piRNA detection method, difficulty in piRNA extraction and detection, etc., and achieve the effect of facilitating centrifugal separation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0026] A method for detecting piRNA in gastric juice, comprising the following main steps in sequence:
[0027]a. Gastric juice extraction and neutralization: extract two fasting gastric juice samples (different parts) with a gastric tube, 5ml each, put them into a centrifuge tube, put them on ice, and then add 0.5% sodium hydroxide solution with a mass percentage concentration of 20%. ml, placed in a shaker at room temperature and shaken for 20 minutes to liquefy the mucus in gastric juice, adjust the pH value to 7.0±0.5 with 20% sodium hydroxide solution by mass percentage, and then add 1% Trypsin (commercially available) solution 20μl, vortex to mix, place in a 37℃ water bath for 15 minutes to digest, hydrolyze the protein in the gastric juice and homogenize the gastric juice, centrifuge at 1000rpm for 20 minutes at 4℃;
[0028] b. RNA release: Take 200 μl of the supernatant from step a and place it in a sterile centrifuge tube, add 50 μl of sterilized DEPC (commercially av...
Embodiment 2
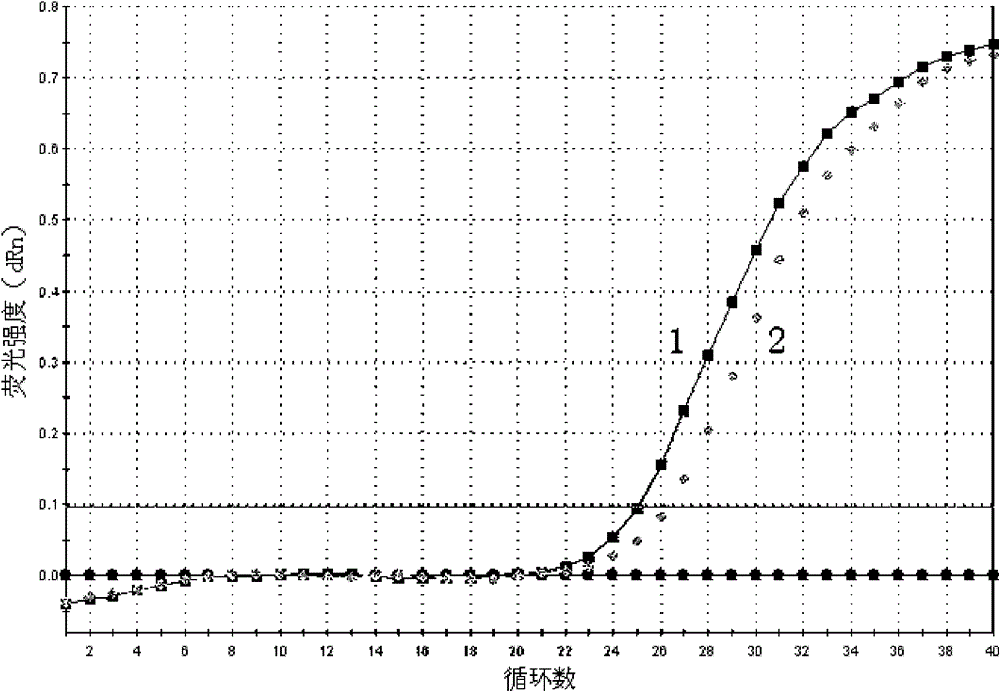
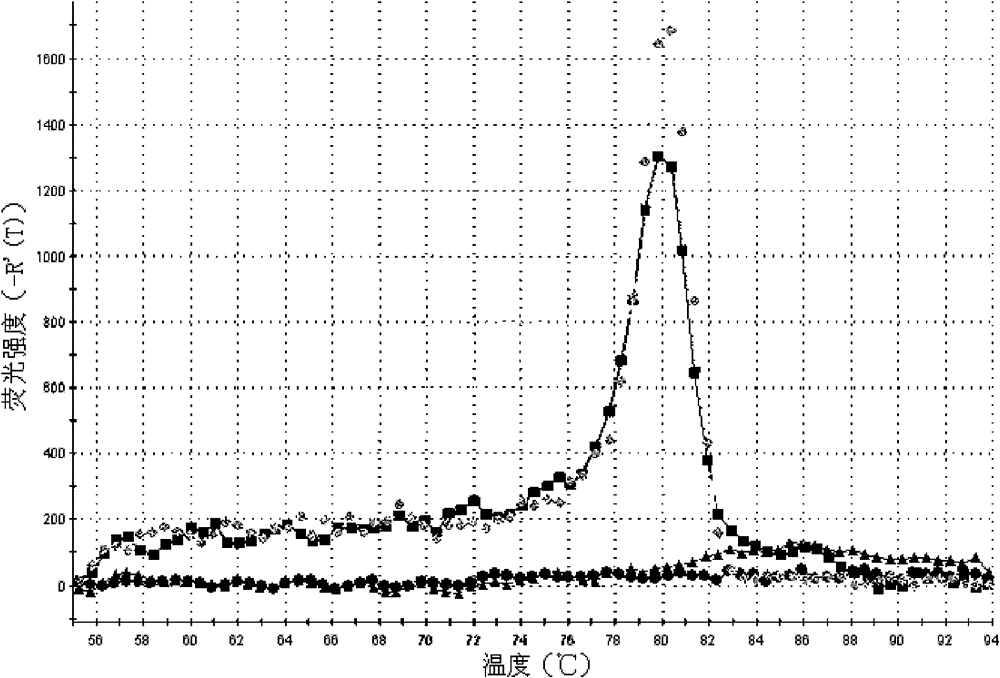
[0035] The method is basically the same as in Example 1, except that in step g, the amplified upstream primer is replaced by the piR-823 specific amplification upstream primer for piR-651, and the amplification curve of the expression level of piR-823 in gastric juice is detected (attached image 3 ) and melting curve (with Figure 4 ), from the amplification curve, the Ct values of the two gastric juice samples detected are 22.27 and 22.91 respectively; from the melting curve, it can be known that the curve is a narrow single peak, and it can be seen that the piR of each gastric juice sample -823 were amplified specifically without the interference of primer dimers and heterogeneous bands.
Embodiment 3
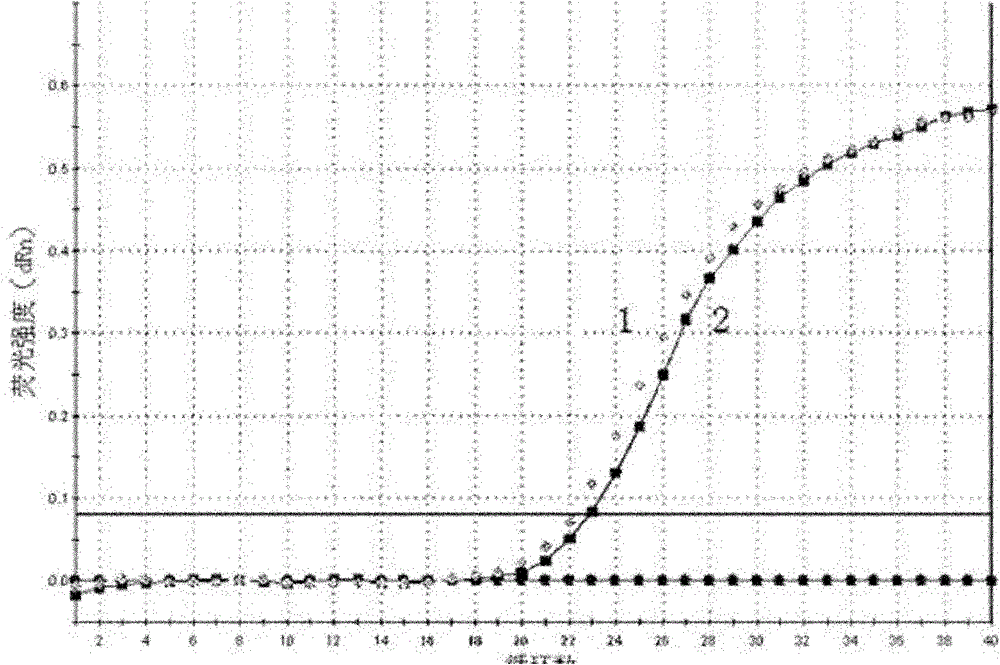
[0037] The method is basically the same as in Example 1, except that in step g, the amplified upstream primer is replaced by the small RNA U6 specific amplification upstream primer for piR-651, and the amplification curve of the expression level of small RNA U6 in the gastric juice is detected ( attached Figure 5 ) and melting curve (with Image 6 ), from the amplification curve, the Ct values of the two gastric juice samples detected are 29.60 and 32.04 respectively; from the melting curve, it can be known that the curve is a narrow single peak, and it can be seen that each gastric juice sample has a small RNA U6 was amplified specifically without the interference of primer dimers and heterogeneous bands.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com