Multi-sample mixed sequencing method and kit
A hybrid sequencing and kit technology, used in biochemical equipment and methods, combinatorial chemistry, organic compound libraries, etc., can solve problems such as low mixing uniformity, and achieve the effects of increasing uniformity, shortening operation time, and simplifying steps.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0035] Example 1: Preparation of the second-generation high-throughput sequencing library
[0036] The following is a brief description of the preparation steps of the second-generation multi-sample hybrid sequencing library:
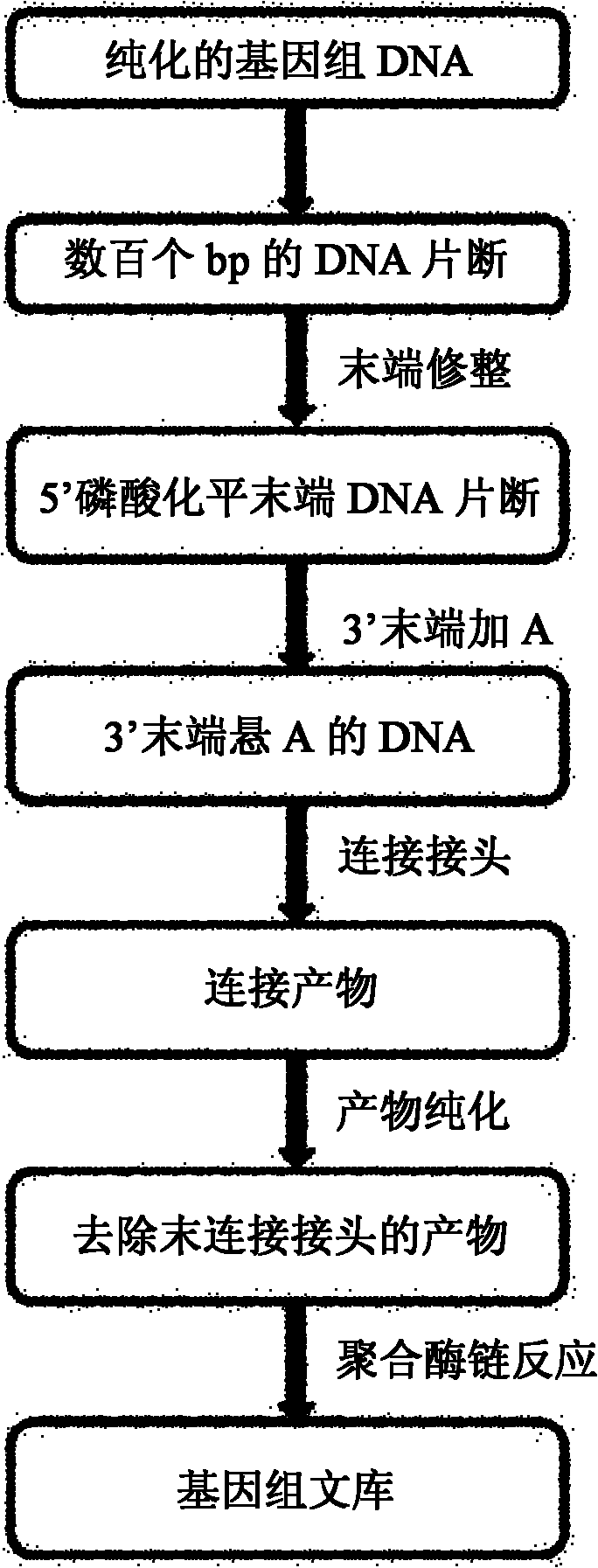
[0037] DNA fragments that break to a specified range;
[0038] End filling and 5' end phosphorylation: jointly completed by the enzyme Klenow (New England Biolabs), T4 phosphorylase and DNA polymerase, and then the product was cleaned and purified;
[0039] Overhang A at the end: under the action of klenow ex-(New England Biolabs) (an improved Klenow enzyme, its 3'-5'exocutting activity is lost), the product of the previous step is overhanging the A base at the end of the double strand, The product is then cleaned and purified;
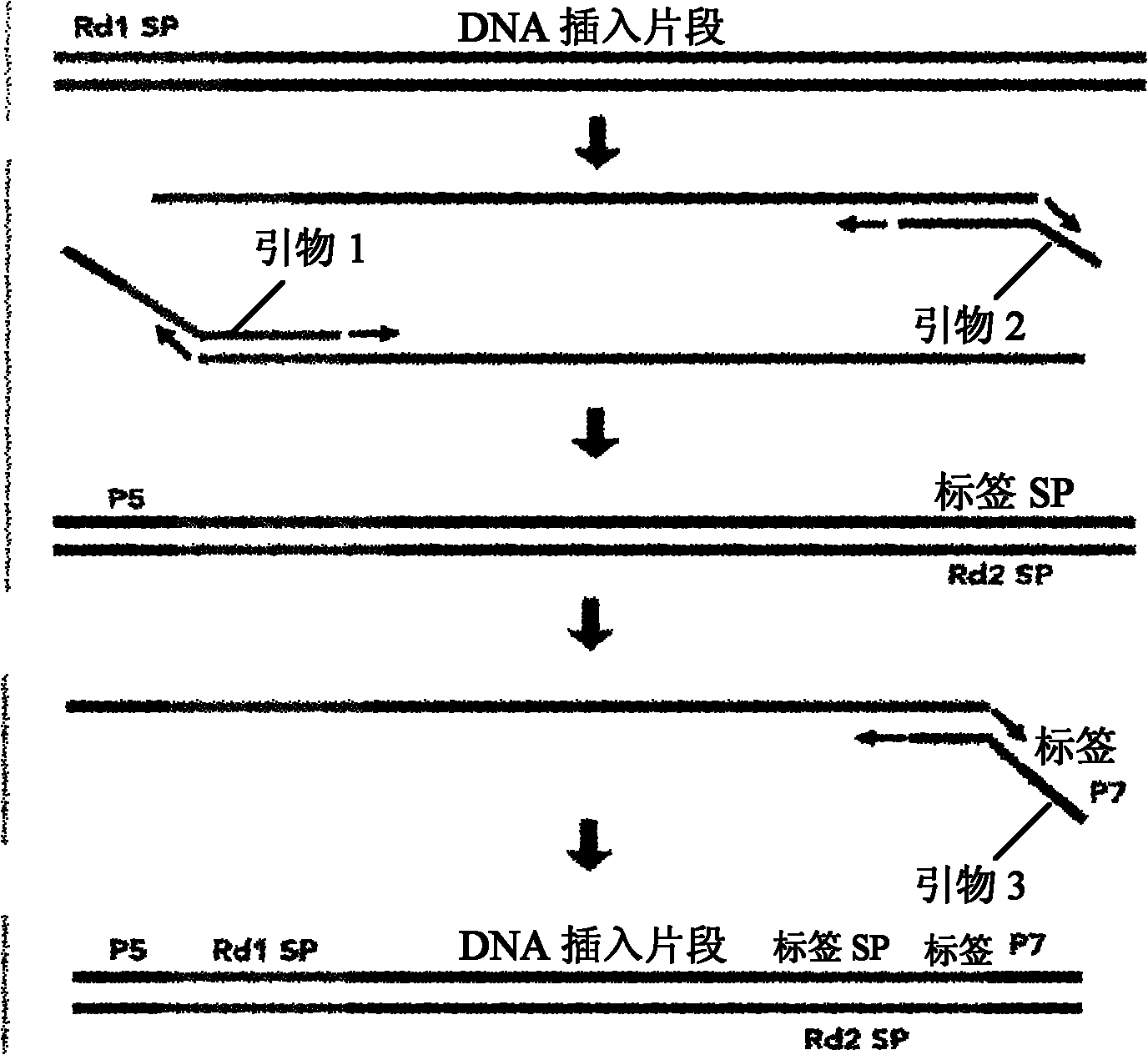
[0040] Connection: According to the different sources of samples and the planned channels to connect different adapters, T4DNA ligase is required, and different adapters have different tag sequences in Table 5 to distinguish ...
Embodiment 2
[0095]Embodiment 2: Kit for preparing multi-sample DNA mixed sequencing library
[0096] The kit for preparing a multi-sample DNA mixed sequencing library provided by the present invention mainly includes the following components:
[0097] Enzymes for repairing fragmented DNA ends and enzymes for 5' phosphorylation of DNA ends, including enzymes Klenow, T4 phosphorylase and DNA polymerase, as well as buffers required by various enzymes;
[0098] Enzymes that add adenine to the 3' end of DNA, such as klenow ex-(3'-5'exocutting activity loss), and dATP;
[0099] An enzyme such as DNA ligase that ligates the Y-linker to DNA;
[0100] Buffers suitable for various enzymes such as DNA ligase buffer, Klenow enzyme buffer and T4 DNA ligase buffer (NEB);
[0101] Y-type adapter mixture, including a variety of Y-type adapters, the tag sequences included in each Y-type adapter are different, such as various tag sequences listed in Table 5, including a Y-type adapter pair of a tag seque...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com