Method for rapid detection of drug resistant gene New Delhi metallo-beta-lactamase in bacteria
A technology of lactamase and drug-resistant genes, applied in the biological field, to achieve the effect of eliminating non-specific amplification
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
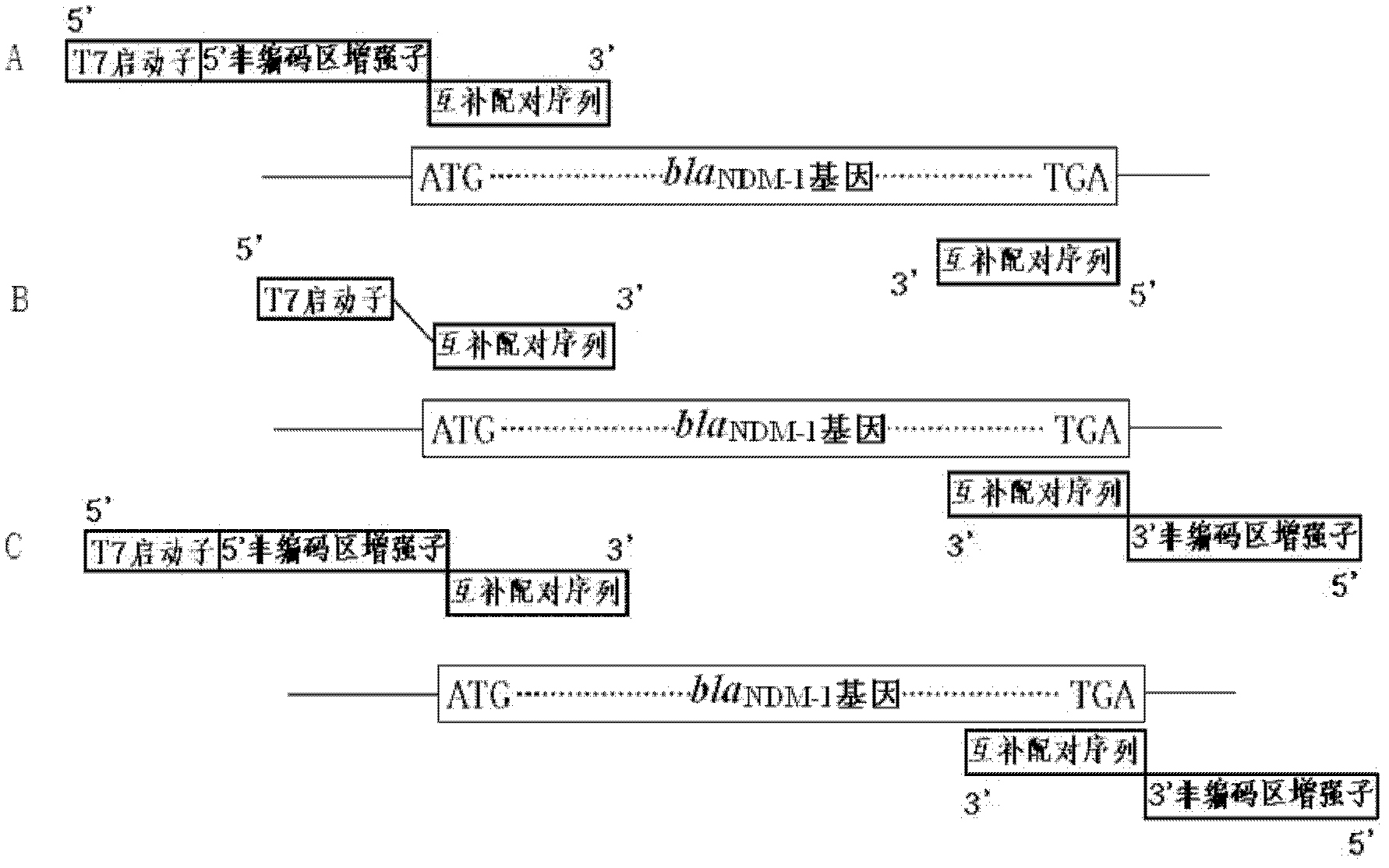
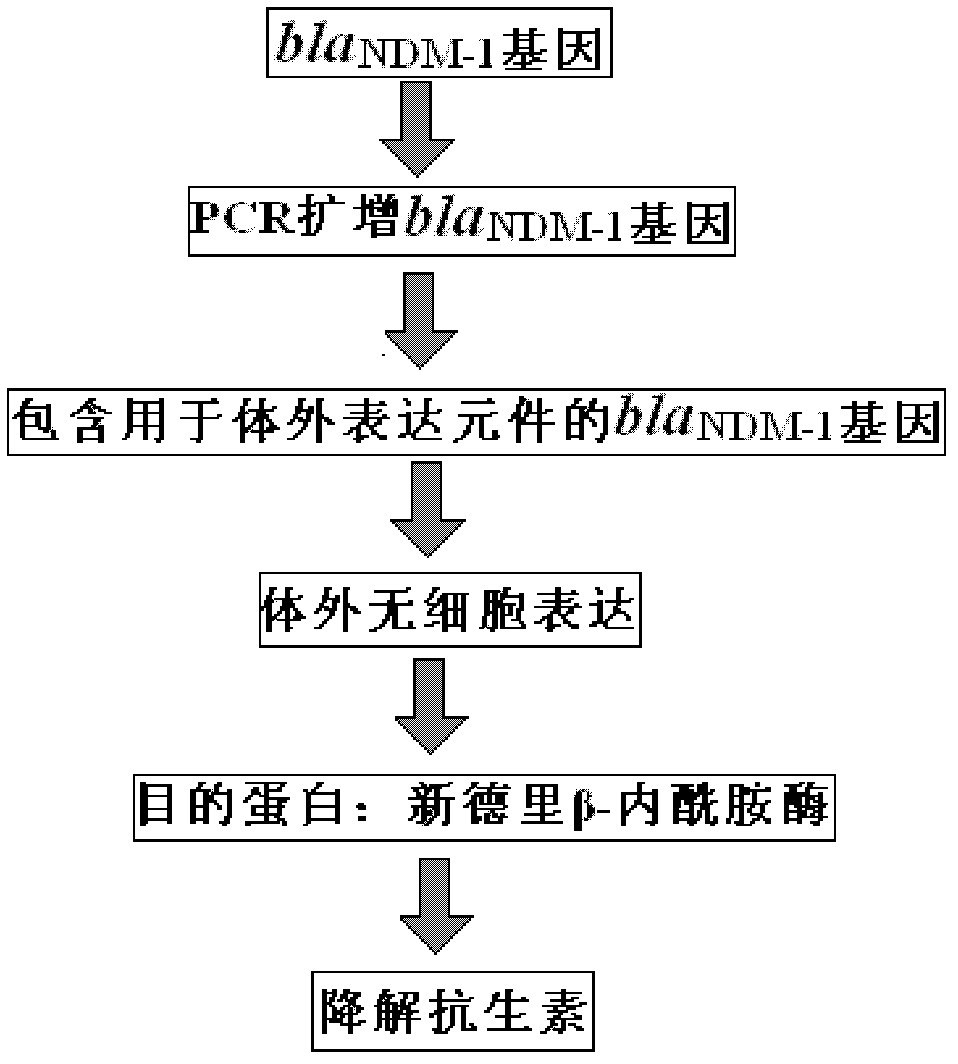
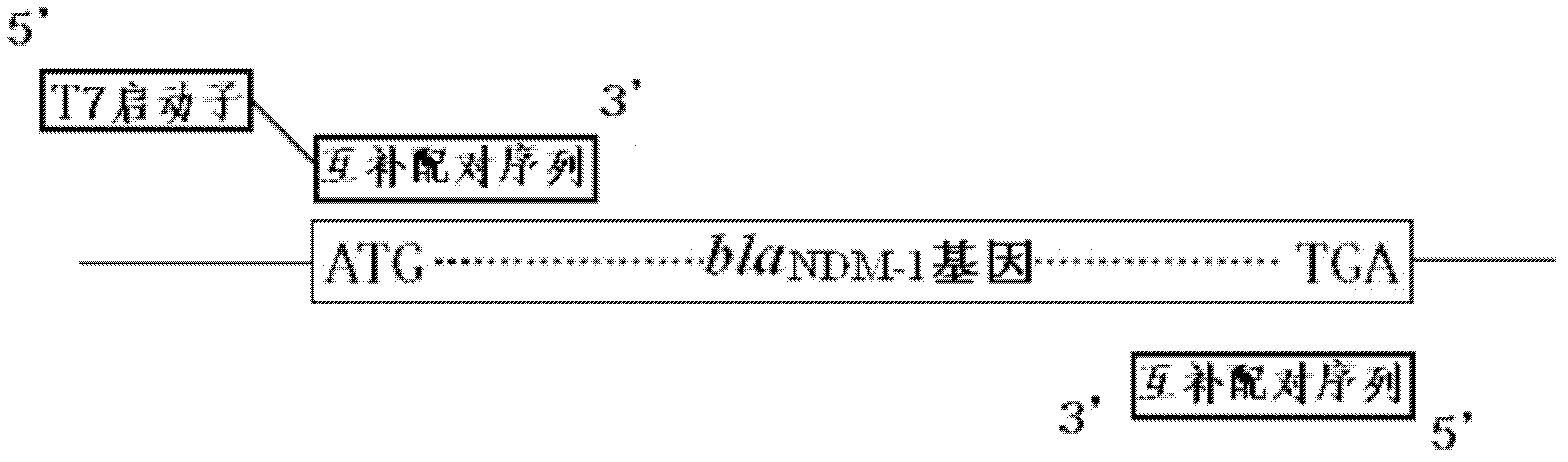
[0035] A rapid assay for the detection and characterization of the drug-resistant gene New Delhi β-lactamase in bacteria (full-length amplified by PCR containing the T7 promoter bla NDM-1 gene for detection of β-lactamase resistance gene and characterization of its encoded enzyme in New Delhi), the steps are:
[0036] 1. Boiling and lysing Mycobacterium tuberculosis to quickly prepare templates for PCR reactions:
[0037] Take a small amount (about 100 – 10000 bacteria) containing bla NDM-1 Genetic strains (e.g. containing bla NDM-1 Genetic Escherichia coli, Klebsiella pneumoniae and Klebsiella stinky nose, etc.) add 100ul double distilled water to boil for 10min, then centrifuge at 13000rpm for 5min, and carefully draw the supernatant as amplification bla NDM-1 Gene template.
[0038] 2. PCR amplification bla NDM-1 Gene primer design. Upstream primer sequence P1: TAATACGACTCACTATAGGATGGAATTGCCCAATATTATGCA 3', downstream primer sequence: 5' TCAGCGCAGCTTGTCGGCCATGC 3'...
Embodiment 2
[0059] Full-length PCR amplification with T7 promoter and 5' UTR enhancer bla NDM-1 Genes used in the detection of β-lactamase resistance genes and characterization of their encoded enzymes in New Delhi:
[0060] 1. Boiling and lysing Mycobacterium tuberculosis to quickly prepare templates for PCR reactions:
[0061] Take a small amount (about 100 – 10000 bacteria) containing bla NDM-1 Genetic strains (e.g. containing bla NDM-1 Genetic Escherichia coli, Klebsiella pneumoniae and Klebsiella stinky nose, etc.) add 100ul double distilled water to boil for 10min, then centrifuge at 13000rpm for 5min, and carefully draw the supernatant as amplification bla NDM-1 Gene template.
[0062] 2. PCR amplification bla NDM-1 Gene primer design. Upstream primer sequence P2: 5' TAATACGACTCACTATAGGATACTCCCCCACAACAGCTTACAATACTCCCCACACAGCTTACAAATACTCCCCATGGAATTGCCCAATATTATGCA 3';
[0063] Downstream primer sequence: 5'TCAGCGCAGCTTGTCGGCCATGC3'.
[0064] PCR amplification conditions: ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com