Sirt4 and uses thereof
A technology of fatty acid oxidation and compound, applied in the field of SIRT4 and its use, can solve the problem that the role is not described
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
[0112] Preparation and screening of combinatorial chemical libraries is well known to those skilled in the art. Such combinatorial chemical libraries include, but are not limited to, peptide libraries (see, e.g., U.S. Patent 5,010,175; Furka, Int. J. Pept. Prot. Res. 37:487-493 (1991) and Houghton et al., Nature 354:84-88 (1991)). Other chemistries for generating chemical diversity libraries can also be used. Such chemicals include, but are not limited to: peptoids (e.g., PCT Publication No. WO 91 / 19735), encoded peptides (e.g., PCT Publication No. WO 93 / 20242), random biooligomers (e.g., PCT Publication No. WO 92 / 00091), benzodiazepines class (e.g., U.S. Patent No. 5,288,514), diversomers such as hydantoins, benzodiazepines Peptides and dipeptides (Hobbs et al., Proc. Nat. Acad. Sci. USA 90:6909-6913 (1993)), ethylene-like peptides (Hagihara et al., J Amer. Chem. Soc. 114:6568 (1992) ), non-peptide peptidomimetics with glucose scaffolds (Hirschmann et al., J Amer. Chem...
Embodiment 1
[0194] Example 1: SIRT4 is downregulated during fasting.
[0195] To investigate whether modulation of SIRT4 activity plays a role in response to nutrient deprivation in the liver, livers of fasted 129 / Sv mice were analyzed by quantitative RT-PCR as described above. SIRT4 gene expression. The fasting period was initiated at the beginning of the light cycle (9AM) and food deprivation continued for 24 hours. At the onset of the dark cycle (the period when mice start eating normally), SIRT4 The level was downregulated by 20% (t=10 hours), and after 24 hours of fasting, compared with the starting feeding level of SIRT4, SIRT4 Transcript levels were reduced by half (pfigure 1 A). Because SIRT3 and SIRT5 are also mitochondrial NAD-dependent sirtuins that may be involved in the redundant regulation of hepatic metabolism, the expression of SIRT3 and SIRT5 was also examined by quantitative RT-PCR. In contrast to the downregulation of SIRT4 after fasting, nutrient deprivation ind...
Embodiment 2
[0196] Example 2: Loss of SIRT4 enhances lipid catabolism gene expression following nutrient deprivation.
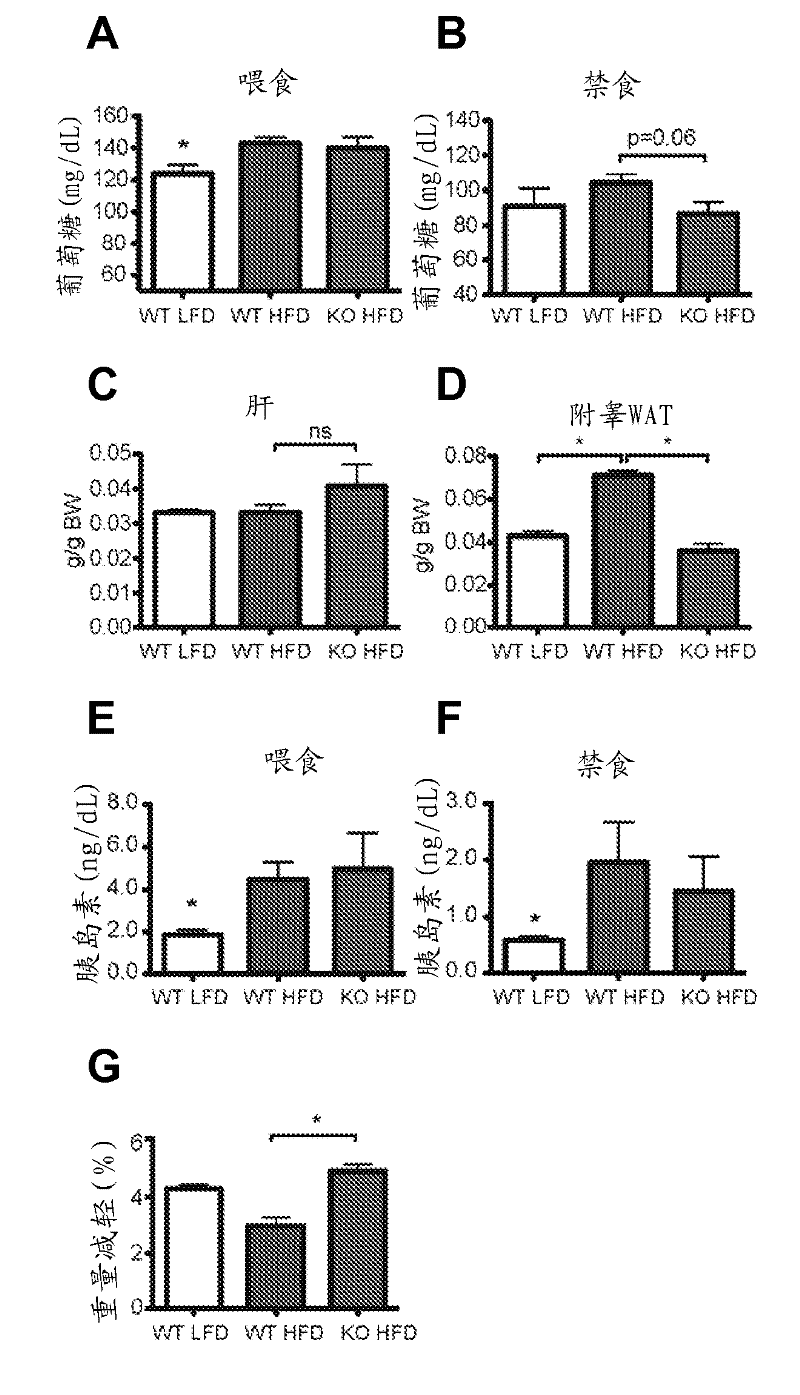
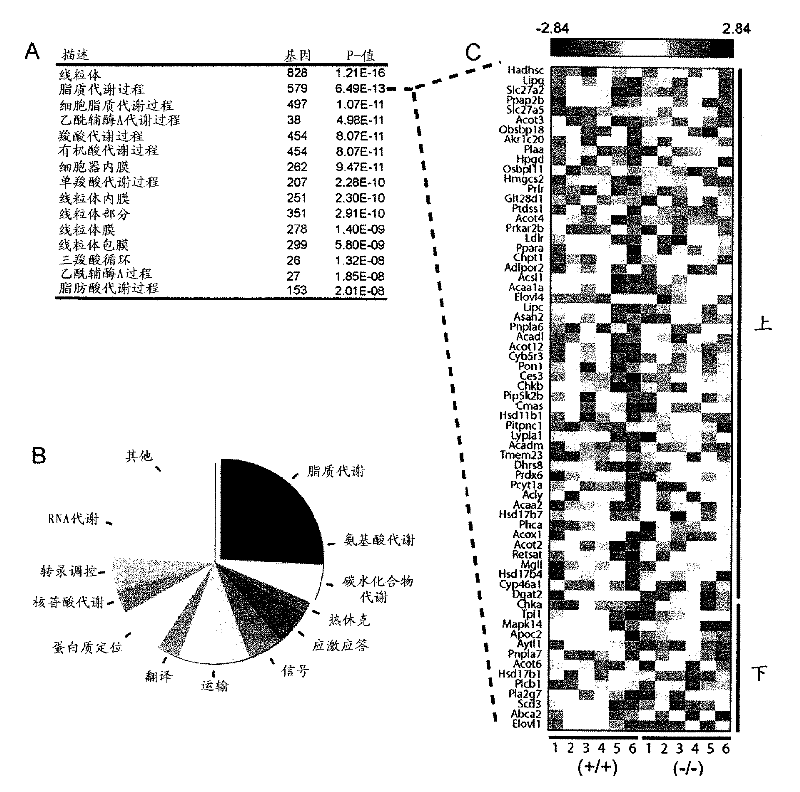
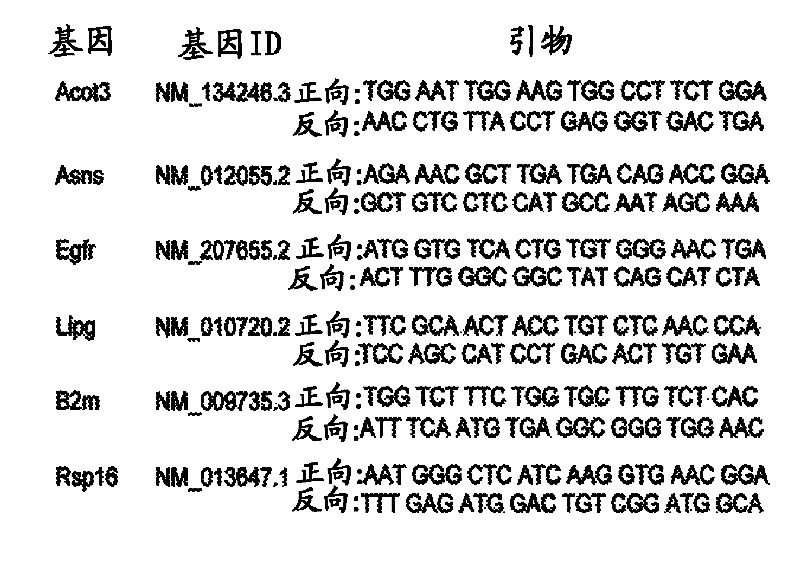
[0197] To characterize the physiological pathways regulated by SIRT4 in the liver after fasting, SIRT4 knockout (KO) and SIRT4 wild-type (WT) mouse livers from 16-h fasted mice were analyzed by microarray analysis as described above. Genome-wide gene expression profiles. SIRT4 KO mice are developmentally normal with no overt hepatic phenotype (Haigis et al., (2006) Cell 126, 941-954). Data analysis revealed that the hepatic gene expression profiles of SIRT4 KO mice (n=6) differed only subtly from those of SIRT4 WT mice (n=6). Of the 22,094 unique genes on the microarray, only 654 genes were significantly different (p figure 2 A). In addition, metabolic pathways including lipid, acetyl-CoA, and tricarboxylic acid metabolism were highly enriched ( figure 2 A). These data strengthen further support that SIRT4 is involved in the regulation of hepatic metabolic programs ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com