A method for detecting the single nucleotide polymorphism of cattle fgf21 gene
A single nucleotide polymorphism, FGF21 technology, applied in the direction of biochemical equipment and methods, microbial measurement/inspection, etc., can solve blank and other problems
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
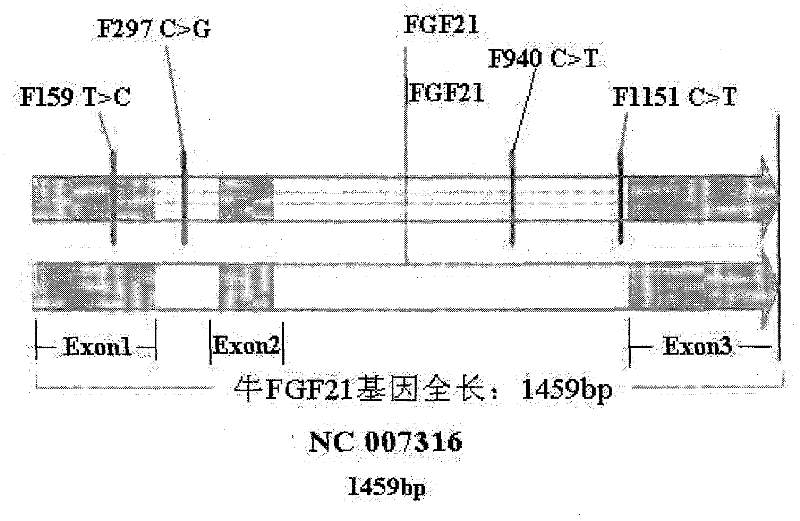
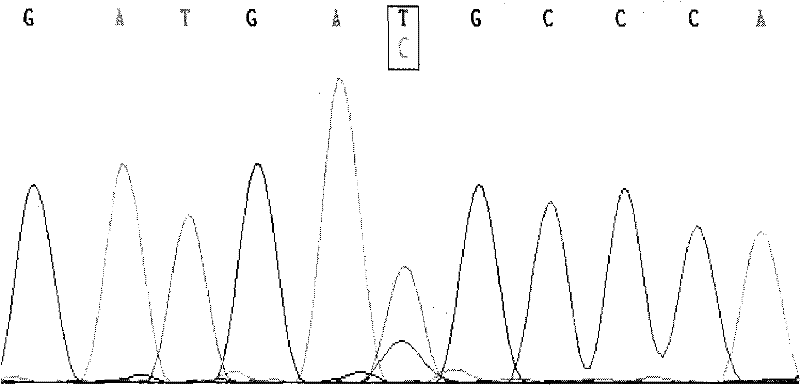
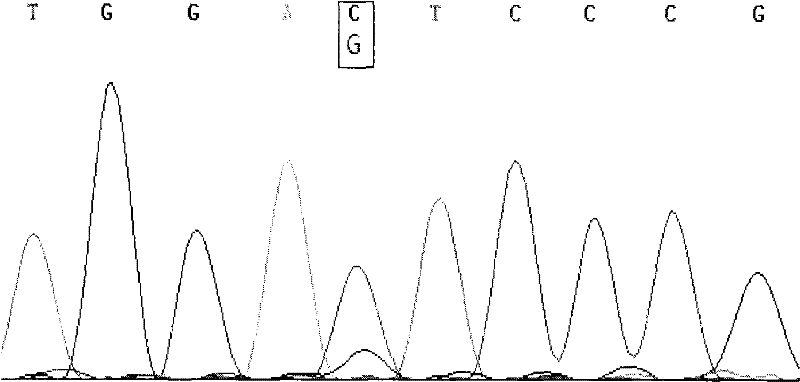
[0046] The present invention utilizes the PCR-RFLP method to detect the single nucleotide polymorphisms at the 159th, 297th, 940th, and 1151st positions of the cattle FGF21 gene. by way of explanation of the invention, not limitation.
[0047] A, design the PCR primers of cattle FGF21 gene
[0048] The full length of cattle FGF21 gene is 1459bp, including 3 exons and 2 introns. Using the bovine (NC_007316) sequence published by NCBI as a reference, use Primer 5.0 to design 3 pairs of primers to amplify the full length of the FGF21 gene (including 3 exons and 2 introns, a total of 11459bp) in 3 segments. The primers are as follows :
[0049] The primer pair P1 is:
[0050] Upstream primer: 5′ATGGGCTGGGACGAGGCCAAGTTC 3′,
[0051] Downstream primer: 5′CAAACCAAGCCTGACCAACATCAAA 3′;
[0052] The primer pair P2 is:
[0053] Upstream primer: 5′GGAAGCTGTACGGATCGGTGAG 3′,
[0054] Downstream primer: 5'CTCCTTTCTCAGCTTTATCGTCTAGG 3';
[0055] The primer pair P3 is:
[0056] Upst...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com