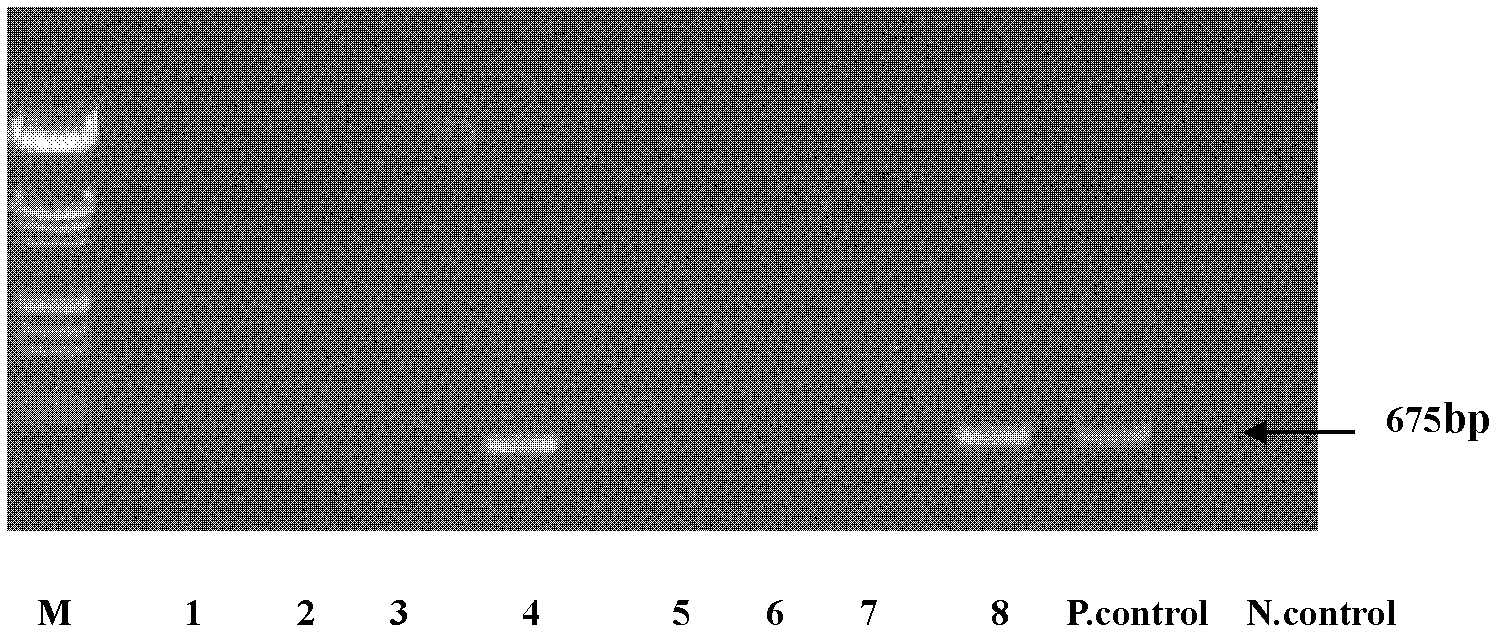Method for identifying Bacillus subtilis for producing lipopeptide surfactants
A technology for producing lipids and lipopeptides, which can be used in biochemical equipment and methods, microbial determination/inspection, DNA/RNA fragments, etc., and can solve problems such as low work efficiency, inability to distinguish lipopeptide microorganisms, and time-consuming efficiency. Achieve the effect of simple operation, short time consumption, high sensitivity and resolution
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
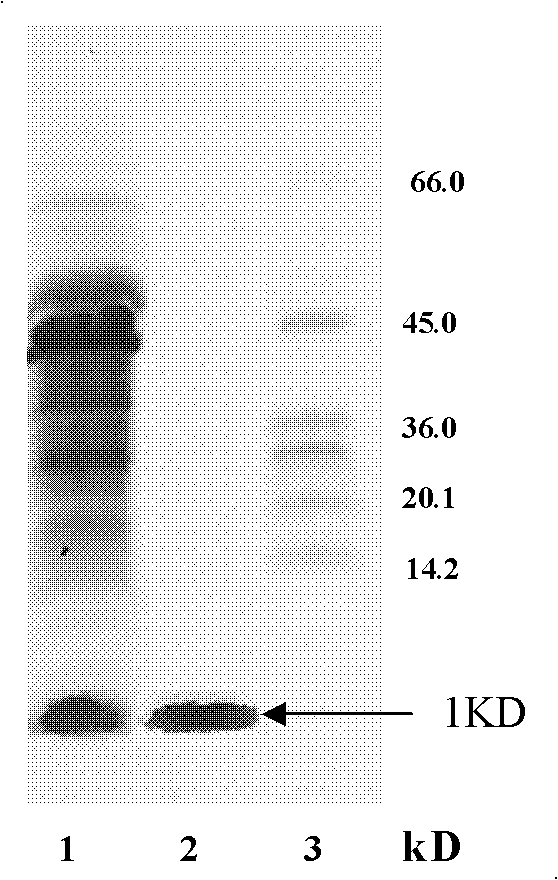
[0037] Embodiment 1, identify the bacillus subtilis that produces lipopeptide surfactant
[0038] 1. Extract bacterial genomic DNA and use it as a template to amplify the srf gene by PCR
[0039]1. Extract bacterial genomic DNA
[0040] Bacterial genomic DNA was extracted by micro-extraction, cells were lysed by SDS, protein was degraded by protease, and polysaccharide components were removed by CTAB. The specific method is as follows:
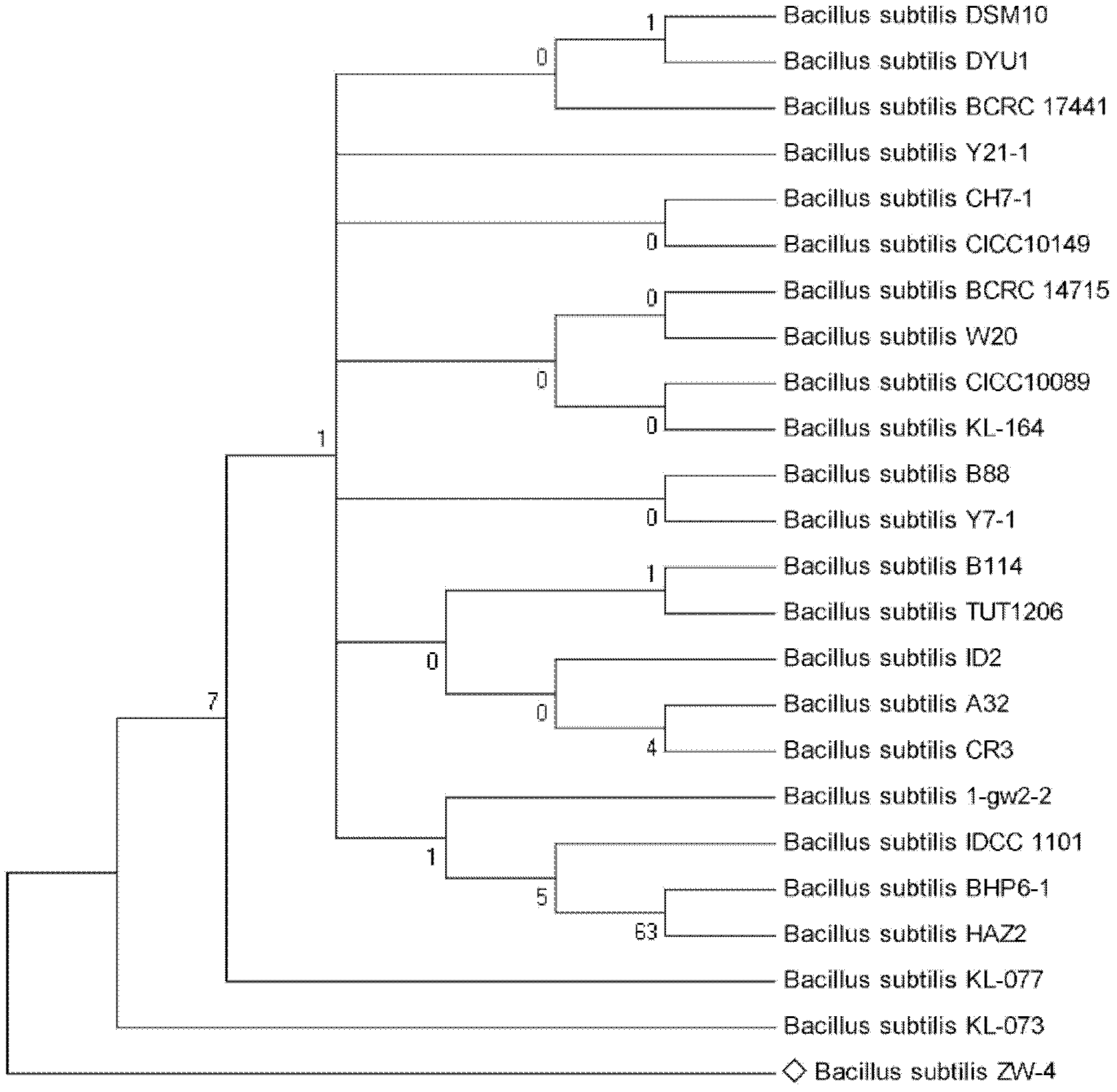
[0041] Strain ZW-1 (Bacillus subtilis ATCC 7058), strain ZW-2 (Bacillus subtilis ATCC21228), strain ZW-3 (Bacillus licheniformis ATCC 9789), strain ZW-4 (Bacillus subtilis ATCC 21770), strain ZW-5 (Bacillus megaterium ATCC 14581), strain ZW-6 (Bacillus pumilus ATCC 7061), strain ZW-7 (Bacillus licheniformis ATCC 14580) and strain ZW-8 (Bacillus subtilis ATCC 15841) (the above strains were purchased from the American Type Culture Collection ( ATCC)) were inoculated in LB medium and cultured with shaking at 180r / min at 37°C for 3 days until t...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com