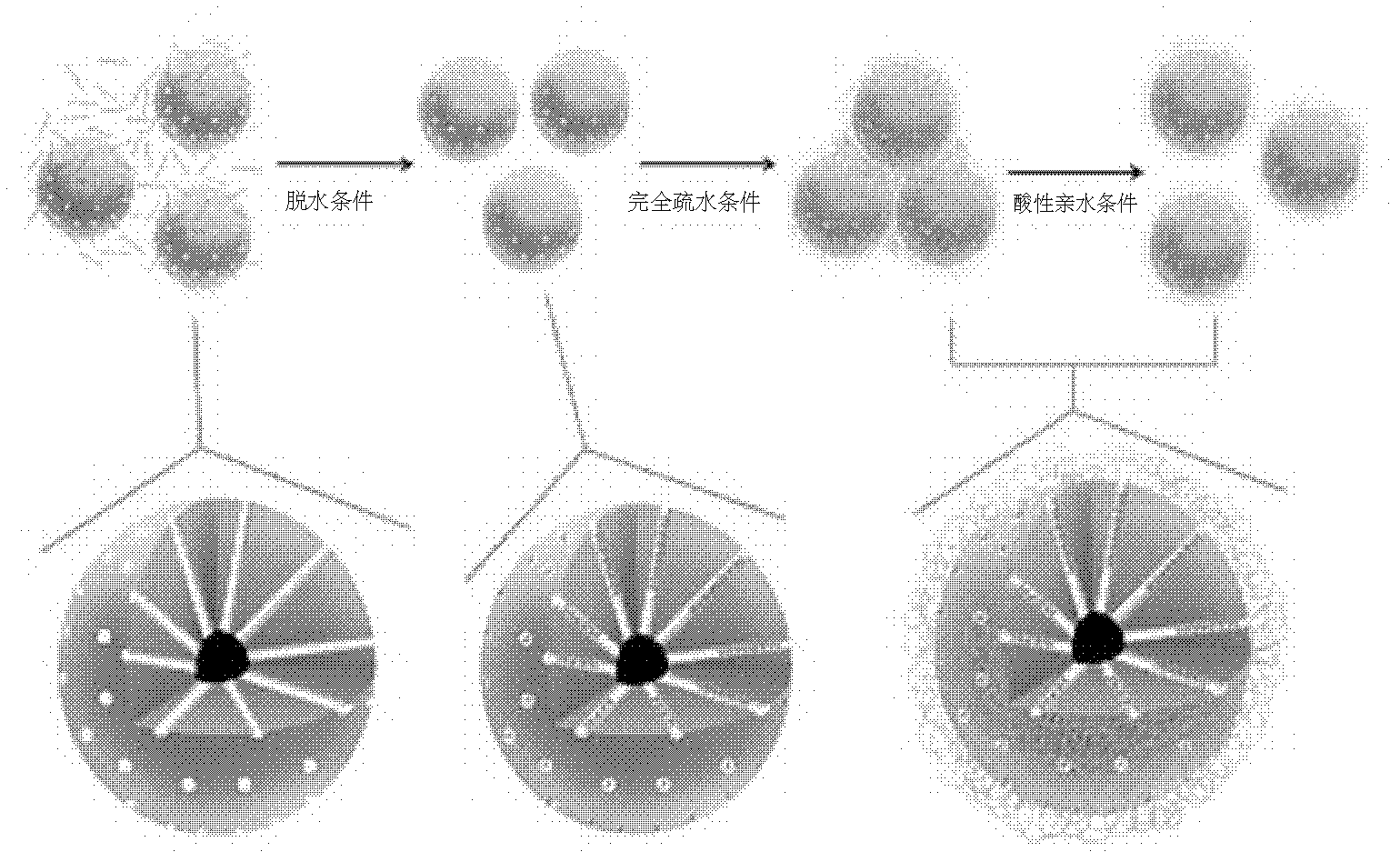
Method for loading siRNA (small interfering Ribonucleic Acid) by using mesoporous silicon dioxide nanoparticles
A technology of mesoporous silica and nanoparticles, which is applied in the direction of pharmaceutical formulations, medical preparations of non-active ingredients, gene therapy, etc., can solve the problems of high specific surface area and failure to exert the channel structure of mesoporous nanoparticles, and achieve Avoid chemical modification, easy-to-handle effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example 1
[0026] 1. First add 0.2mg of M-MSNs into a 2mL centrifuge tube, then add 0.03mL of pure ethanol to it, and ultrasonically oscillate for 30s at 100w to disperse the particles evenly in the solution.
[0027] 2. Add 0.01 mL of high-salt solution (the composition of this high-salt solution is: 0.04 mmol of guanidine hydrochloride, 0.0025 mL of isopropanol, 0.0075 mL of sterilized water, pH 5) to the final solution obtained in step 1 and mix uniform.
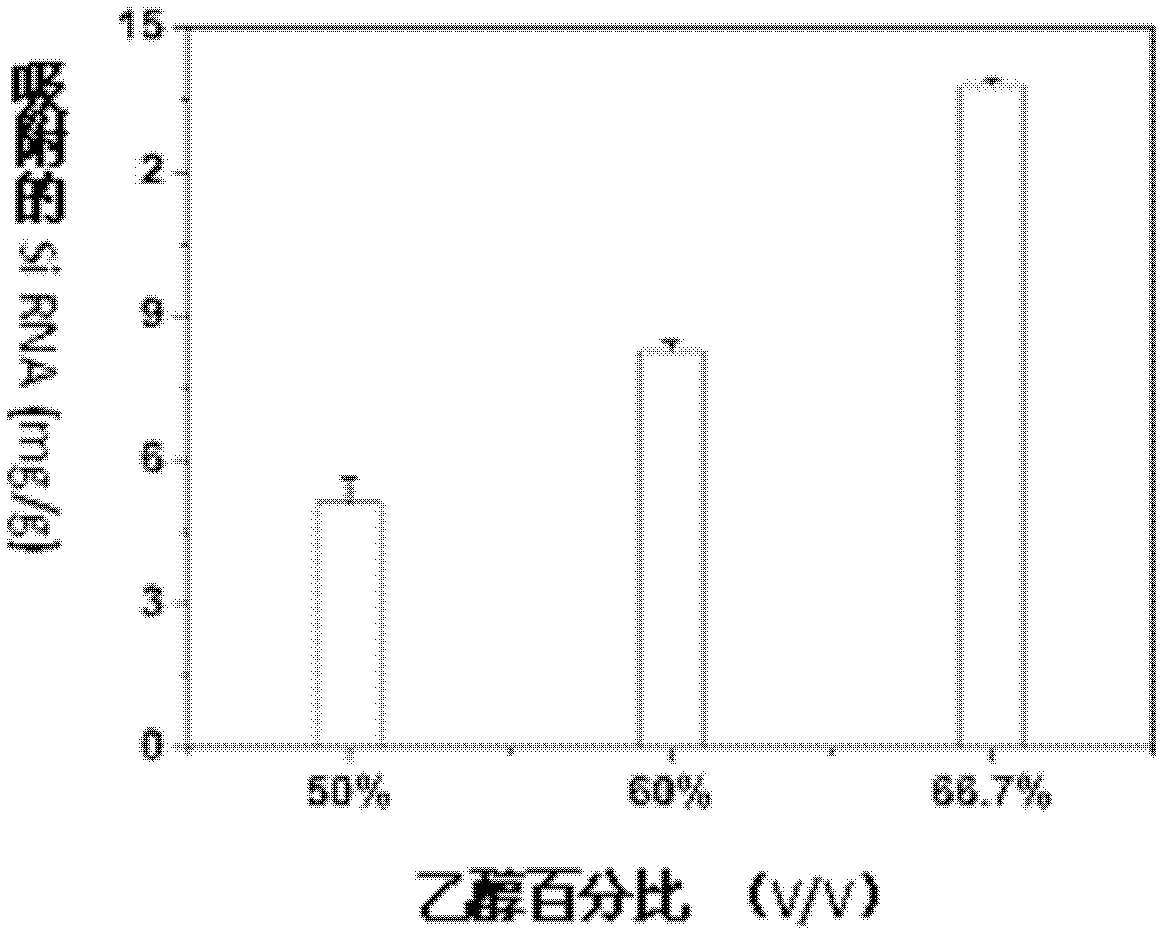
[0028]3. Add 0.01 mL of siRNA aqueous solution (solvent is deionized water) with a concentration of 0.6 mg / mL to the solution obtained in step 2, and mix well again. In this embodiment, ethanol: high salt solution: siRNA aqueous solution three The volume ratio between the two was 3:1:1, the mixture was placed in an oven at 25°C for 4 hours, and the M-MSNs adsorbed by siRNA were separated from the solution by centrifugation at 10,000 rpm. At this time, the unit loading capacity of M-MSNs to siRNA was 8.8mg / g ( figure 2 shown).
...
example 2
[0034] 1. Keep the other conditions of steps 1 to 3 in Example 1 unchanged, change the volume ratio of ethanol: high salt solution: siRNA aqueous solution (solvent is double distilled water) to 2: 1: 1 or 4: 1 : 1.
[0035] 2. After the above-mentioned finally obtained particles are separated from the adsorption solution, the unit loading capacity of M-MSNs to siRNA is respectively: when the ratio is 2:1:1, the loading amount is 5.3 mg / g, and the ratio is 4: 1:1 loading capacity is 14.5mg / g ( figure 2 shown).
[0036] 3. Wash the separated particles once with 0.06mL pure ethanol, then add 0.06mL pure ethanol, and use 100w ultrasonic vibration to disperse the particles evenly.
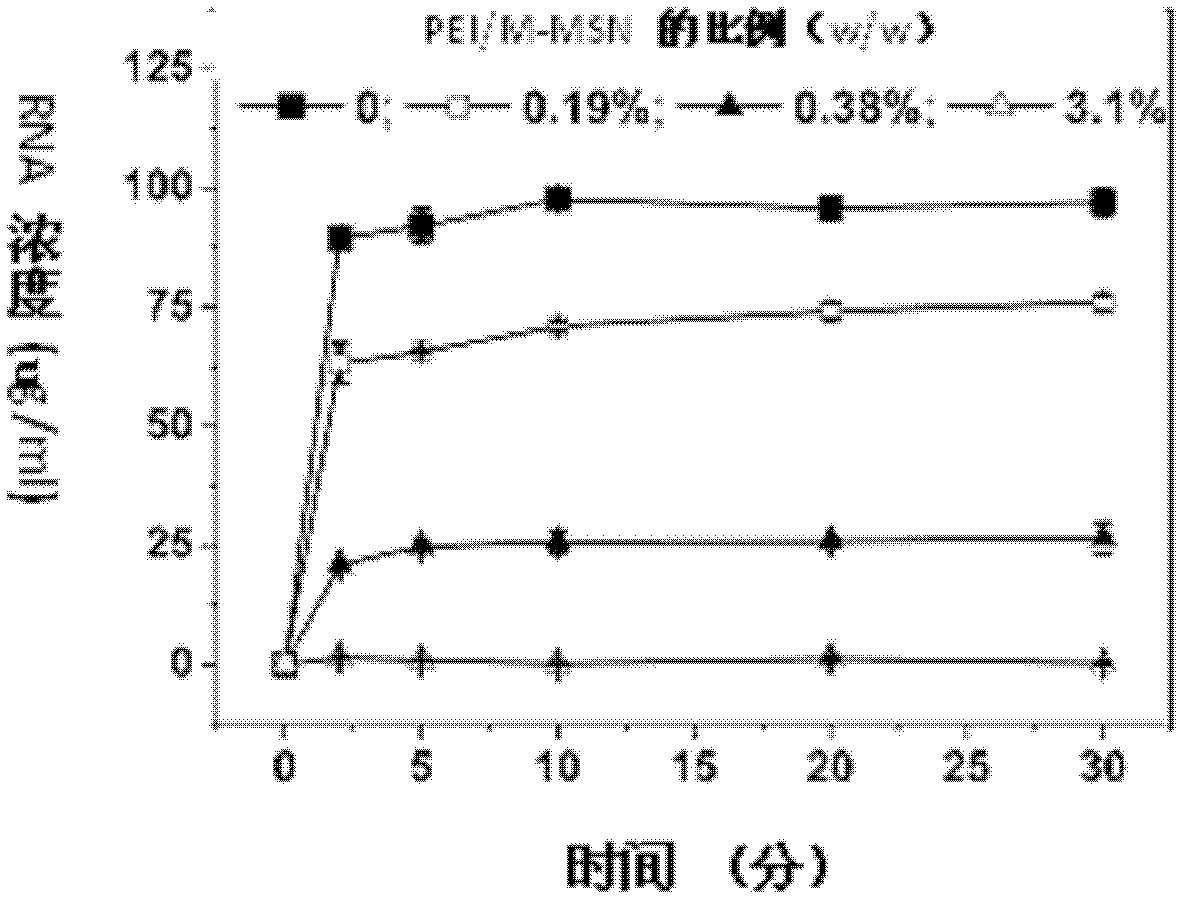
[0037] 4. Add 0.06mL of pure ethanol solution containing polyethyleneimine to the final solution obtained in step 3 (the concentration of polyethyleneimine in pure ethanol solution is 1.5mg / mL), and the mixture is reacted under 100w ultrasonic vibration conditions 20 minutes.
[0038] 5. Separate t...
example 3
[0040] 1. First add 0.2mg of M-MSNs to a 2mL centrifuge tube, then add 0.04mL of pure ethanol to it, and ultrasonically oscillate for 30s at 100w to disperse the particles evenly in the solution.
[0041] 2. Add 0.01 mL of high-salt solution (the composition of this high-salt solution is: 0.04 mmol of guanidine hydrochloride, 0.0025 mL of isopropanol, 0.0075 mL of sterilized water, pH 5) to the final solution obtained in step 1 and mix uniform.
[0042] 3. Add 0.01 mL of siRNA aqueous solution (solvent is sterilized water) with a concentration of 0.5 mg / mL or 2.5 mg / mL to the solution obtained in step 2 and mix the solution evenly. In this embodiment, ethanol: high salt The volume ratio between the solution: siRNA aqueous solution was 4:1:1, the mixture was placed in an oven at 25°C for 4h, and the M-MSNs adsorbed by siRNA were separated from the solution by centrifugation at 10000rpm. The unit loading of siRNA is: when adding the siRNA aqueous solution with a concentration o...
PUM
| Property | Measurement | Unit |
|---|---|---|
| particle diameter | aaaaa | aaaaa |
| pore size | aaaaa | aaaaa |
| molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com