Primer capable of detecting methylation of promoter of gene related to colonic adenocarcinoma
A methylation and gene technology, applied in the determination/inspection of microorganisms, biochemical equipment and methods, DNA/RNA fragments, etc., can solve the problems of limited practical application of detection instruments, cumbersome operation, low sensitivity, etc., to avoid Intra-chain and inter-chain complementarity issues, ensuring specificity, and reducing the effect of background interference
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0083] Embodiment 1, the design of primer
[0084] 1. Selection of candidate genes
[0085] Selected candidate genes with high frequency of methylation associated with colon adenocarcinoma, selected VIM (NM_003380), MGMT (XM_003483574), TMEFF2 / HPP1 (NM_016192), ESR1 (NM_000125), APC (NIM_001127511), and MLH1 (NM_000249) gene sequence. The sequences of these genes, including all exons and introns, can be found through the link http: / / www.ensembl.org. The positions of the amplified sequences are as follows:
[0086] serial number
Location
starting base position
NM_003380
VIM
exon 3
-75~+105
XM_003483574
MGMT
first exon
-370~-224
NM_016192
TMEFF2 / HPP1
first exon
-385~-213
NM_000125
ESR1
exon 6
+62~+226
+NIM_001127511
APCs
first exon
-140~-86
NM_000249
MLH1
first exon
-500~-207
[0087] 2. Design primers
[0088] Find...
Embodiment 2
[0093] Embodiment 2, the application of primer
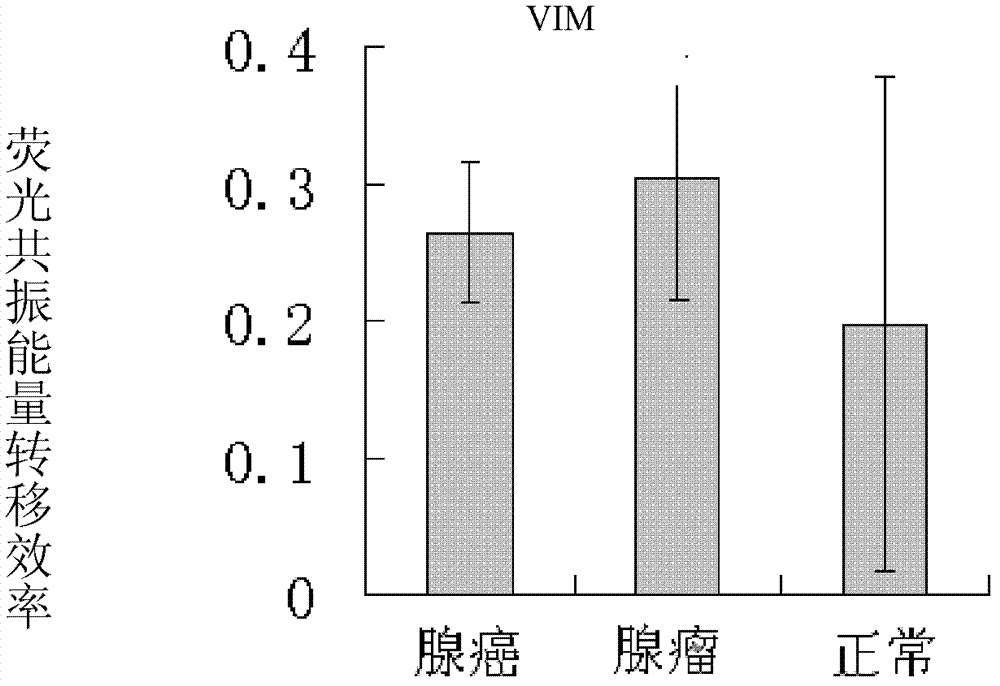
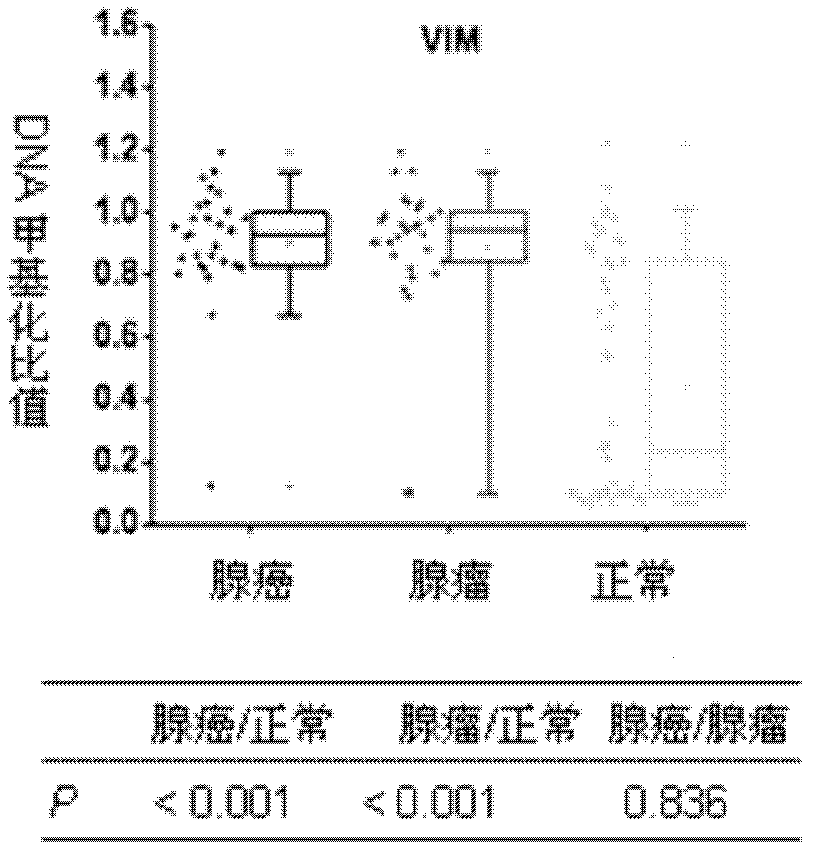
[0094] 1. Application of VIM primers
[0095] The efficiencies of these primers were verified on DNA samples derived from colon adenocarcinoma, adenoma, and normal human tissue samples according to the CCP-FRET detection method reported in references. Specific process:
[0096] 1. Synthesis of PFP
[0097] PFP was synthesized according to the method reported in the reference (Liu, B., Wang, S., Bazan, G.C. & Mikhaillovsky,, A Shape-adaptablewater-soluble conjugated polymers.J.Am.Chem.Soc.2003, 125, 13306-13307) (the structural formula is as above).
[0098] 2. Extraction of DNA to be tested
[0099] Genomic DNA was extracted from 30 colon adenocarcinomas, 30 colon adenomas, and 50 normal human tissue samples (ex vivo blood, from the First Affiliated Hospital of the General Hospital of the Chinese People’s Liberation Army, where the patients were informed and diagnosed).
[0100] The DNA samples obtained above were digested ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com