Primer pair for assisting in identifying Fusarium oxysporum f. sp. phaseoli and application of primer pair
A technology of Fusarium oxysporum and specific primer pairs, which is applied in the field of primer pairs to assist in the identification of the specific type of Fusarium oxysporum, can solve the problems of inability to calculate the content of pathogenic bacteria in kidney bean tissues, and achieve quantitative repeatability, fast, accurate and quantitative Reproducible and reliable results
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0034] Embodiment 1, the preparation of specific primer pair
[0035] The sequence of the specific primer pair (composed of QFopA and QFopB) is as follows:
[0036] QFopA (SEQ ID NO: 1 of the Sequence Listing): 5'-ACATAGCGGTCTACCGTTCG-3';
[0037] QFopB (SEQ ID NO: 2 of the Sequence Listing): 5'-GGTTACAGGAAGCCAAACCA-3'.
[0038] Synthetic QFopA and QFopB.
Embodiment 2
[0039] Embodiment 2, the application (specificity) of specific primer pair in auxiliary identification type of Fusarium oxysporum bean
[0040] The following experiments were carried out on Fusarium oxysporum bean specialization, Fusarium oxysporum chickpea specialization, Fusarium solanisis pea specialization and Phytophthora solani respectively:
[0041] 1. Extract the genomic DNA of the strain.
[0042] 2. Using the genomic DNA in step 1 as a template, perform PCR amplification with the primer pair synthesized in Example 1 to obtain a PCR amplification product.
[0043] The PCR amplification reaction system (20 μL) contains the following components: 100ng genomic DNA, 2mmol L -1 Mg 2+ , 0.25 mmol L -1 dNTPs, 0.25 μmol L -1 QFopA, 0.25 μmol L -1 QFopB, 0.5U Taq DNA polymerase (Dingguo Biotechnology Co., Ltd.).
[0044] The reaction program of PCR amplification: 94°C for 3min; 30 cycles of 94°C for 30s, 55°C for 30s, 72°C for 1min; 72°C for 10min.
[0045] 3. Perfo...
Embodiment 3
[0047] Embodiment 3, the application (sensitivity) of specific primer pair in auxiliary identification type of Fusarium oxysporum bean
[0048] 1. Template preparation
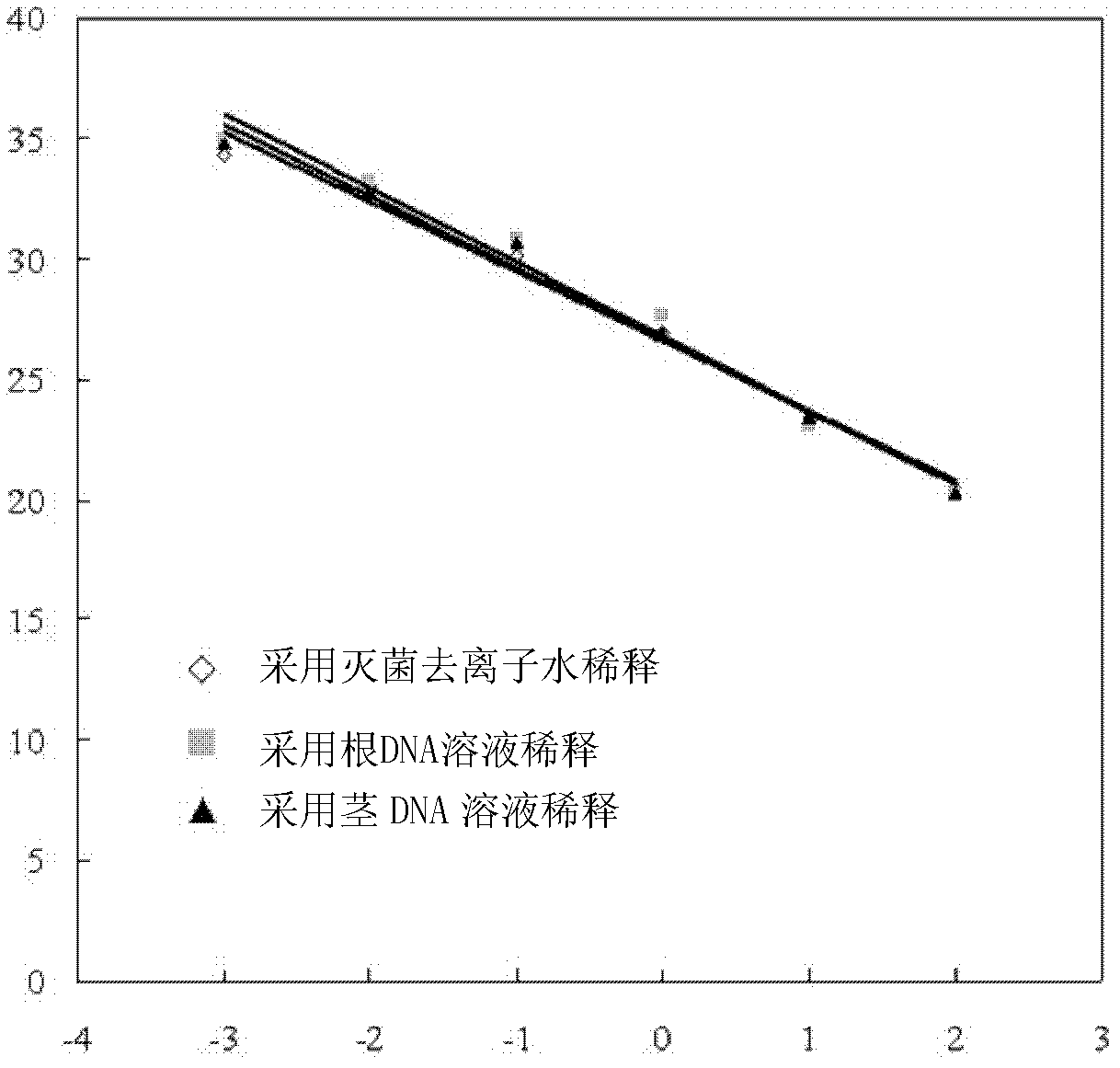
[0049] 1. Extract the genomic DNA of Fusarium oxysporum bean-specialized type, and adjust it to a solution of 50 ng DNA / μL with sterilized deionized water.
[0050] 2. The solution in step 1 was serially diluted to obtain DNA concentrations of 50, 5, 5×10 -1 , 5×10 -2 , 5×10 -3 , 5×10 -4 ng μL -1 The six dilutions (in order of dilution 1 to dilution 6). Sterilized deionized water, root DNA solution and stem DNA solution were used to carry out the gradient dilution respectively, and eighteen kinds of dilutions were obtained in total. The root DNA solution was obtained by extracting genomic DNA from the root of the common bean variety BRB-130, and the DNA concentration was 100 ng / μL. The stem DNA solution is obtained by extracting genomic DNA from the stem of the common bean variety BRB-130, and the DNA c...
PUM
| Property | Measurement | Unit |
|---|---|---|
| PCR efficiency | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com