LncRNA (long noncoding ribonucleic acid) excavating method based on gene sequence expression analysis
A gene sequence, high expression technology, applied in the biological field, can solve the problem that the expression level of long non-coding RNA cannot be determined
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0016] The present invention will take a cancer disease as an example to introduce the specific implementation steps of the present invention
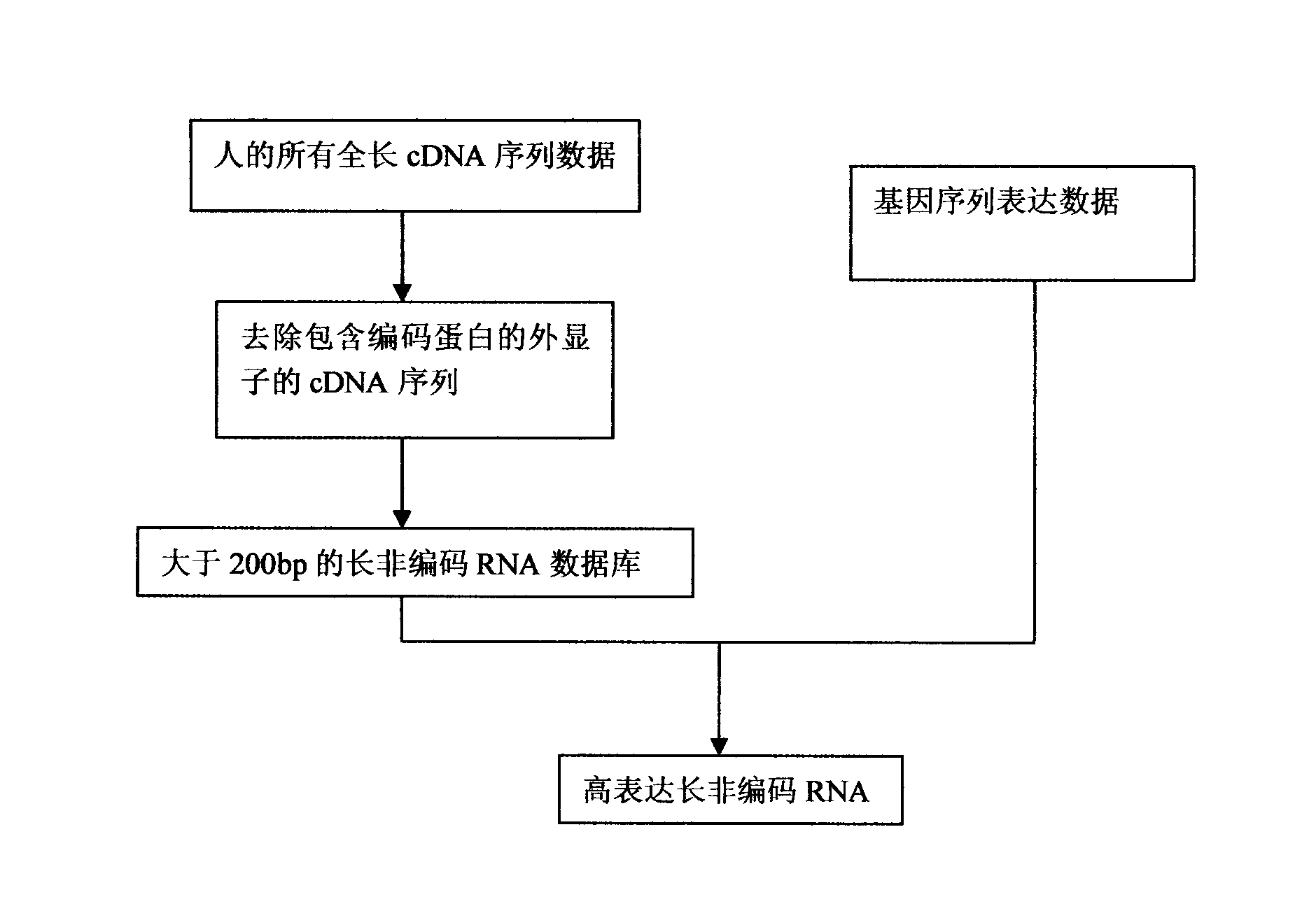
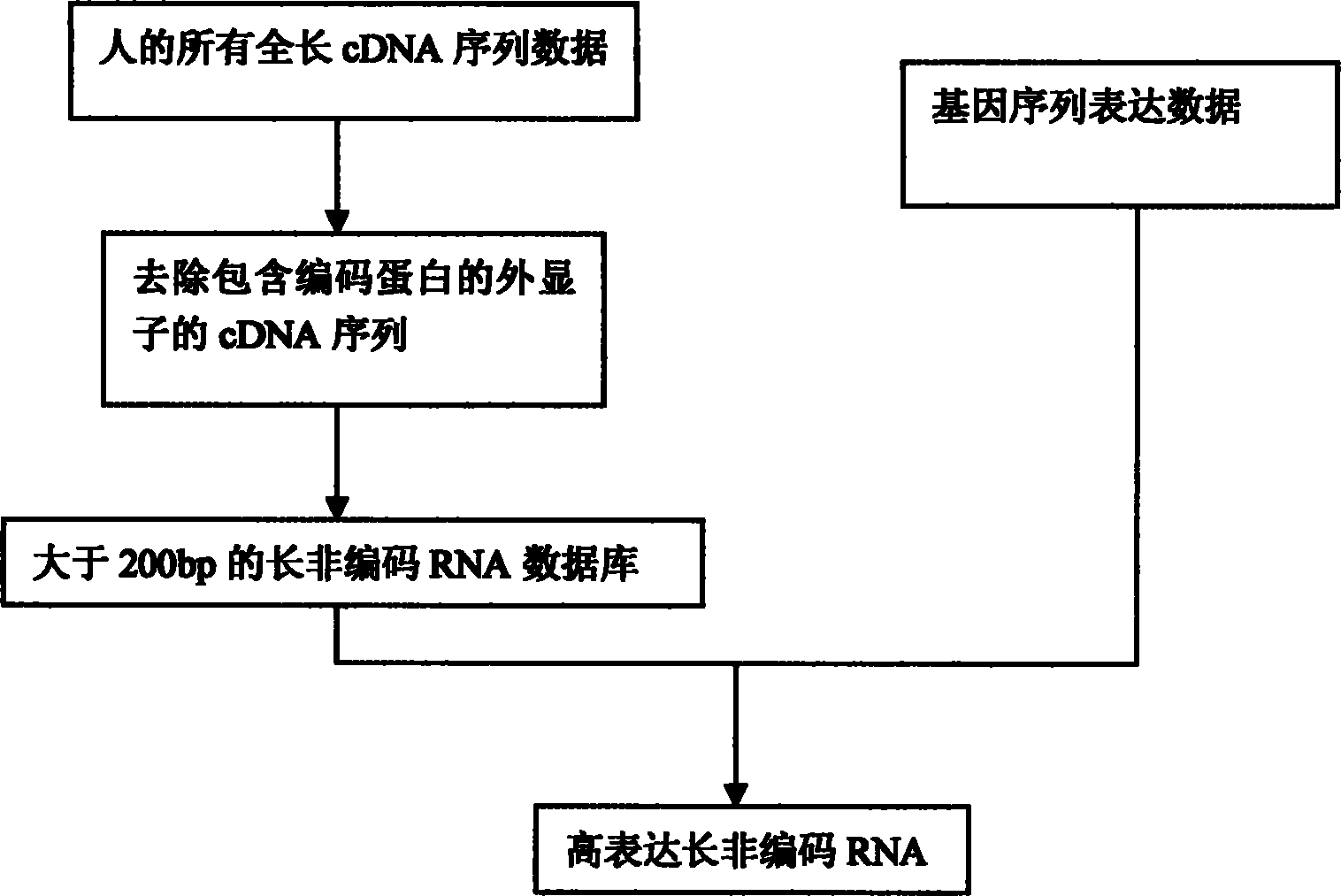
[0017] Step 1: Collect all human full-length mRNA sequence data. The data comes from the NCBI database (http: / / www.ncbi.nlm.nih.gov / nuccore).
[0018] Step 2: Removal of mRNA sequences containing protein-coding exons.
[0019] Step 3: Organize long non-coding RNAs larger than 200bp to form a searchable database.
[0020] Step 4: Search the existing gene expression sequence analysis data to identify highly expressed long non-coding RNAs. We use computer programs to generate virtual digestion results from the long non-coding RNA sequence database. If a restriction does not return a result, the restriction site is automatically added in front of the sequence to ensure the comprehensiveness of the results. The program records important parameters during the entire analysis process, such as the resulting tag sequence of enzyme digestion,...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com