Method for constructing Prunus mume Sieb.et Zucc SSR (simple sequence repeat) genetic map
A technology of genetic maps and construction methods, applied in biochemical equipment and methods, microbial determination/inspection, DNA/RNA fragments, etc. Marking and other issues to achieve the effect of speeding up the process
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0027] The construction of embodiment 1 plum blossom genetic mapping population
[0028] According to the principle of parent selection for mapping, the plum blossom variety 'Fenpetal' with a high seed setting rate and well-developed stigma was used as the female parent, and the plum blossom variety 'Button Yudie' with a large amount of loose pollen and high pollen viability was used as the male parent.
[0029] During the early flowering stage and the full flowering stage of the male parent, choose the flower buds of the big bud and the middle bud stage on the branches without damage by diseases and insect pests and physiological defects, peel off the anthers and spread them on sulfuric acid paper, room temperature (25 ℃) humidity (below 35%), Naturally loose powder for 20 hours and store at 4°C for later use. At the initial flowering stage of the female parent, select the short and medium flowering branches that grow robustly, trim them appropriately, and carefully remove th...
Embodiment 2
[0030] Example 2 Acquisition of the SSR core primer set developed based on the whole genome sequence of Meihua
[0031] 1.1 Development of 670 pairs of SSR primers based on the whole genome sequence of Meihua
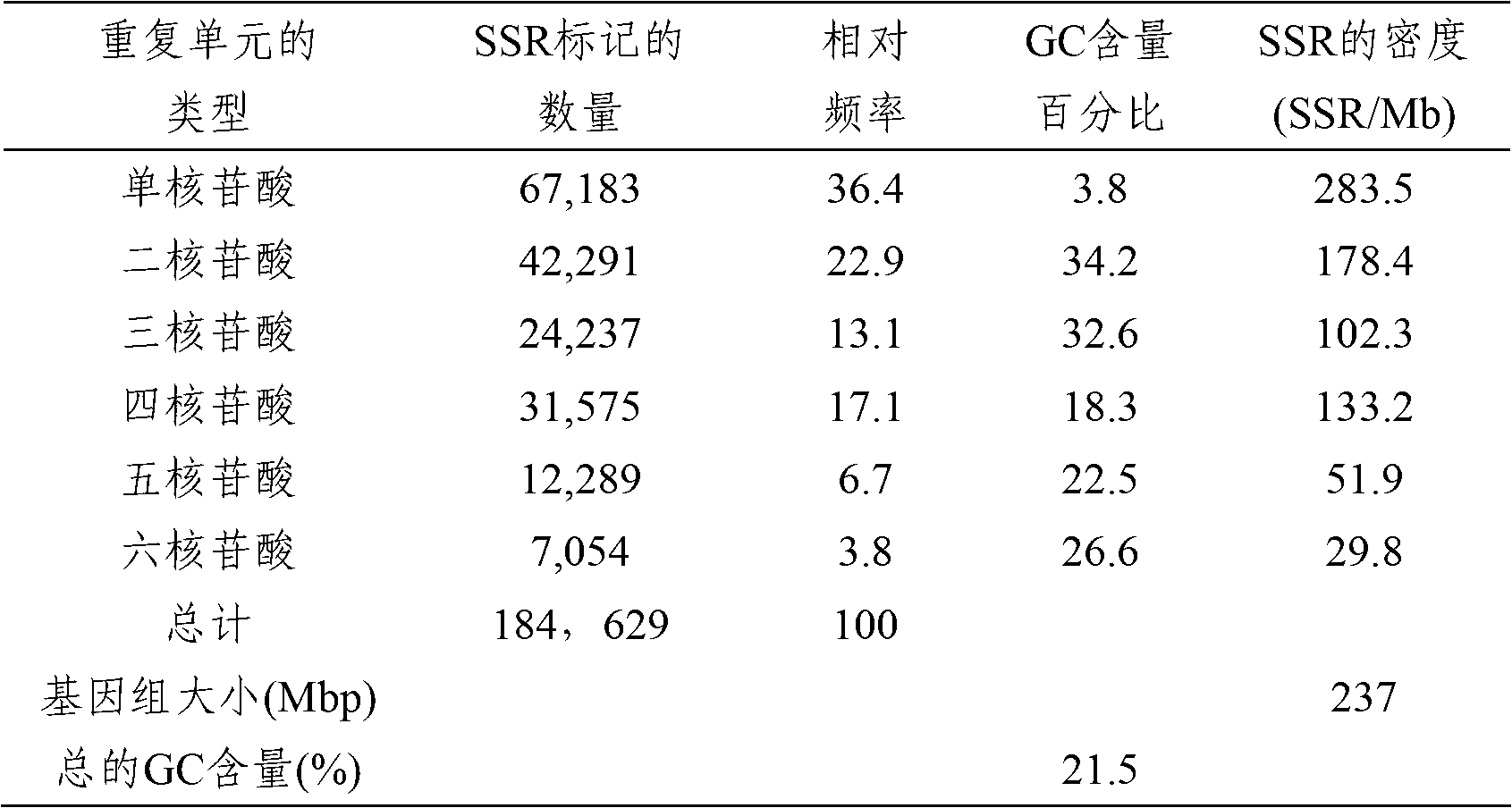
[0032] On the basis of using the Solexa sequencing technology of Illiumina to complete the whole genome sequence of plum blossom, use the computer program MISA (MIcroSAtellites identification tool, http: / / pgrc.ipk-gatersleben.de / misa) to search for 1- For microsatellite markers of 6 nucleotide repeat units, the standards adopted are as follows: the minimum repeat length of a single nucleotide repeat unit is 10 bp, the minimum repeat length of a dinucleotide to tetranucleotide repeat unit is 12 bp, and the minimum repeat length of a pentanucleotide repeat unit is 12 bp. The minimum repeat length of the nucleotide repeat unit is 15 bp, and the minimum repeat length of the hexanucleotide repeat unit is 18 bp. In short, there are 184629 microsatellite markers in the whole g...
Embodiment 3
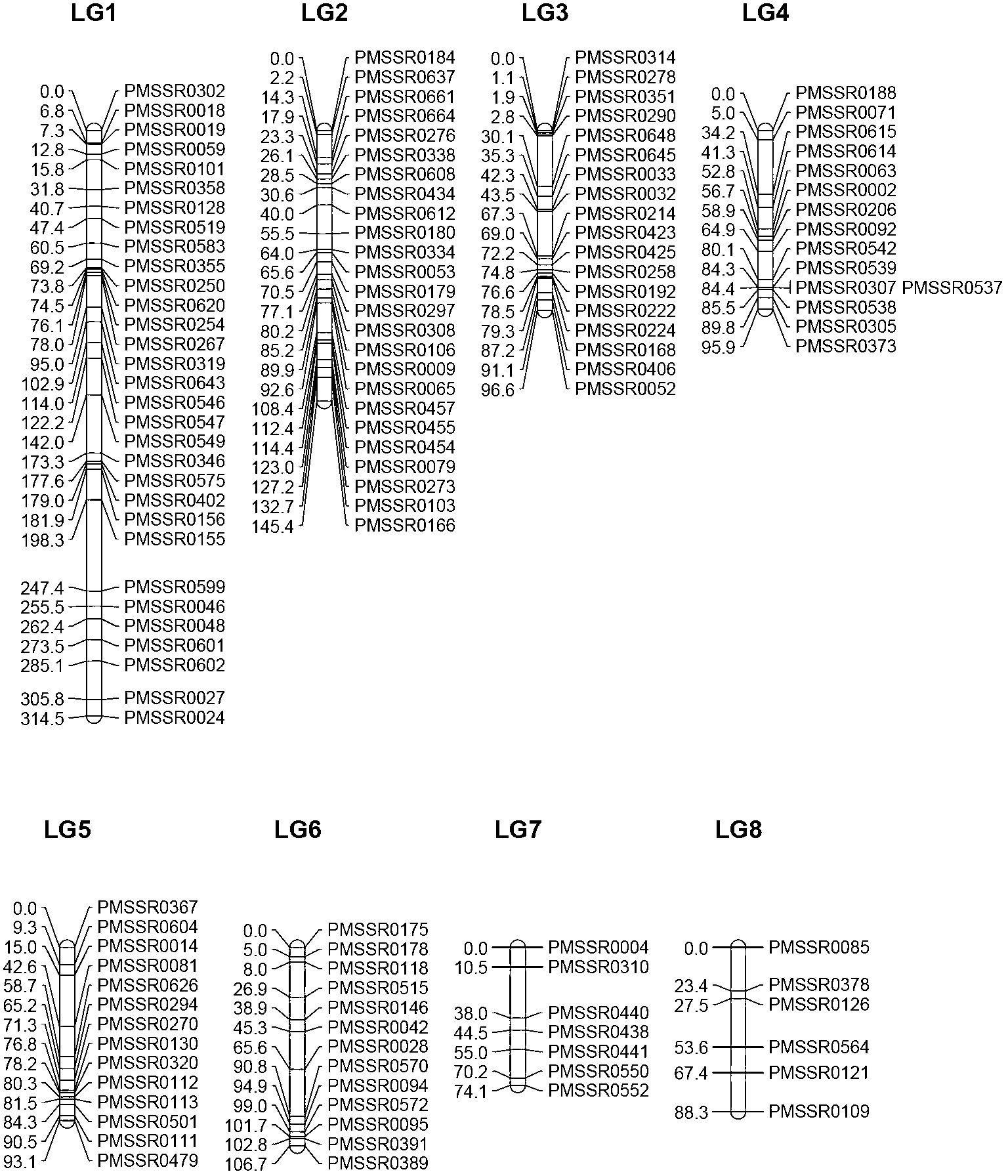
[0043] Example 3 Construction of Meihua SSR Genetic Mapping
[0044] 1.1 Extraction of parental genomic DNA
[0045] From the F1 population obtained in Example 1, 190 trees were randomly selected to construct the SSR genetic map. Take 1-3 young leaves below the parietal leaves of the parents and their progeny, and extract the total DNA of the leaves with the Plant Genome Extraction Kit (DP305-02) of Tiangen Company, so that the DNA concentration reaches 50 ng / μl.
[0046] 1.2 PCR amplification using 144 pairs of SSR core primer sets
[0047] (1) Fluorescently label the upstream primers of the screened 144 pairs of SSR core primer sets with FAM, HEX and TEMRA;
[0048] (2) Using fluorescently labeled SSR primers, the parents and their 190 offspring randomly selected for the construction of genetic maps were amplified by PCR;
[0049] Reaction system (10 μL): 10× buffer (containing Mg 2+ ) 1μL, 2.5mM dNTP 0.8μL, Taq DNA polymerase (Promega, Madison, WI, USA) 1U, 10mmol L -1...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com