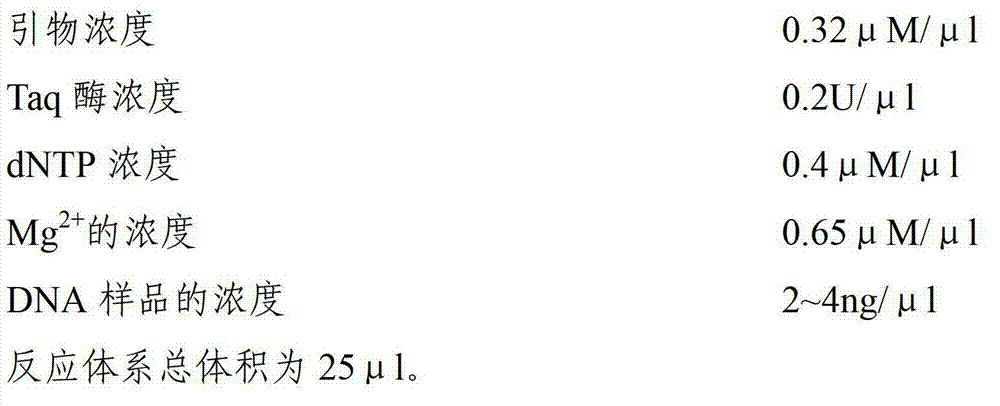Simple sequence repeat (SSR) marker polymerase chain reaction (PCR) reaction method applied to chrysanthemum and related species thereof universally
A technology of labeling, chrysanthemum, applied in the field of plant biology, to achieve the effect of easy operation, good repeatability and good versatility
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0026] Embodiment 1 The establishment of the common SSR marker PCR reaction method of Chrysanthemum and its related species in the present invention
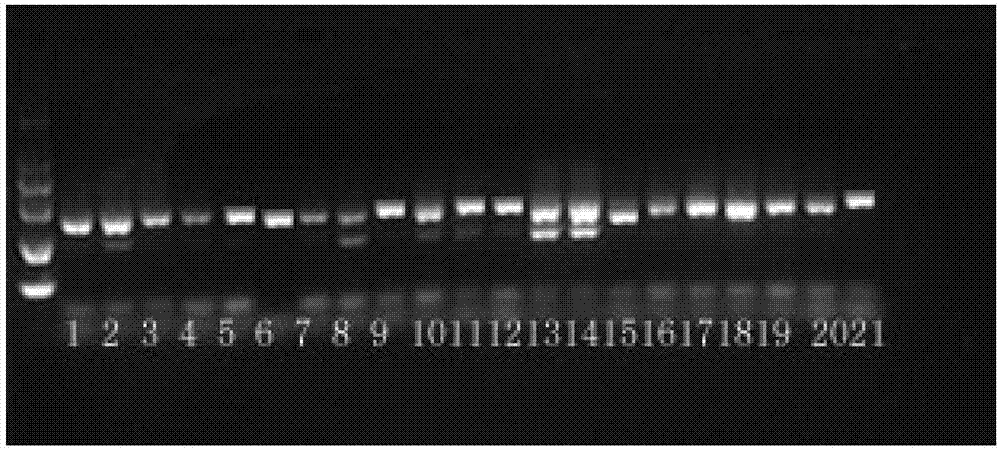
[0027] (1) DNA samples of commonly used wild species and their cultivated varieties in Chrysanthemum, Chrysanthemum, Taihang Chrysanthemum, and Hibiscus were extracted.
[0028] The wild species of chrysanthemum are: wild chrysanthemum, chamomile, chamomile, Maohua chrysanthemum, small red chrysanthemum, chrysanthemum brain, purple wild chrysanthemum, and Shennong chrysanthemum.
[0029] Chrysanthemum cultivars include: Sunshine Chrysanthemum (potted chrysanthemum), Fanbailu (northern ground cover chrysanthemum), Sijihuang (northern ground cover chrysanthemum), Xiangyu (cut flower chrysanthemum), Shanghai chrysanthemum (southern ground cover chrysanthemum), Xiaju (summer flowering variety), Guoqinghong (small chrysanthemum in southern ground cover), Yurenmian (chrysanthemum variety for tea);
[0030] The chrysanthemums in Chrys...
Embodiment 2
[0063] Embodiment 2 utilizes the method of the present invention to carry out the screening of chrysanthemum SSR marker
[0064] (1) According to the design principles of SSR molecular markers, according to the published EST sequence of chrysanthemum, use the Primer5 design software to design 3 pairs of SSR primers:
[0065] SSR marker YH-1 primer sequence:
[0066] Upstream primer: 5'-TTCGCATGGGTGGTGGGTT-3' (as shown in SEQ ID NO.5)
[0067] Downstream primer: 5'-CAACAACACAACATGGGGG-3' (as shown in SEQ ID NO.6)
[0068] SSR marker YH-2 primer sequence:
[0069] Upstream primer: 5'-TTCGACGGGTGGTGGTGTT-3' (as shown in SEQ ID NO.3)
[0070] Downstream primer: 5'-GACACCACAACGCTACCGC-3' (as shown in SEQ ID NO.4)
[0071] SSR marker YH-3 primer sequence:
[0072] Upstream primer: 5'-GGGACCCAAGCAGTAGAATG-3' (as shown in SEQ ID NO.7)
[0073] Downstream primer: 5'-ACCAAACAACCCTCCAACC-3' (as shown in SEQ ID NO.8)
[0074] (2) Extract DNA samples of chrysanthemum varieties Yulon...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com