Assays and kits for serotyping pseudomonas aeruginosa and oligonucleotide sequences useful in such methods and kits
A technology of Pseudomonas aeruginosa and oligonucleotides, which is applied in the field of oligonucleotides and Pseudomonas aeruginosa serotype-specific antibodies, and can solve problems such as high difficulty and inability to achieve detection accuracy.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0141] Example 1: Microbiological samples
[0142] To improve the 4-valent Pseudomonas aeruginosa serotyping test, 16 reference strains and 94 clinical isolates were used. Reference strains were obtained from Institut Pasteur (Paris, France) or Belgium bacteria collection BCCM (Ghent, Belgium). Clinical isolates were obtained from university hospitals in Basel, Bern, Zurich, Switzerland or Jena, Germany, and from multi-national clinical studies on cystic fibrosis.
[0143] To compare a modified 4-valent Pseudomonas aeruginosa serotyping test with a commercial agglutination test, nearly five hundred respiratory Pseudomonas aeruginosa isolates collected from approximately 20 hospitals in the United States, Germany, Greece, and Belgium were used. things.
Embodiment 2
[0144] Embodiment 2: sample preparation
[0145] 2.1 Measurement of purified bacterial DNA
[0146] Each reference strain and clinical isolate was cultured overnight at 37°C in 5 ml of brain heart infusion medium BHI (from BD Bioscience). Bacterial DNA was isolated from bacterial cell pellets using a high-purity PCR template preparation kit (from Roche Diagnostics). DNA stocks of all DNA samples were prepared as 20 ng / μl standards prior to use in PCR. The DNA stock solution was serially diluted 1:10 with ultrapure water (from Roche Diagnostics) as a working solution. Take 5 μl of each DNA working solution as template solution.
[0147] 2.2 Measurements from bacterial cultures
[0148] Reference strains and clinical isolates were plated on BHI agar plates (from BD Bioscience). After overnight incubation at 37°C, one loop of bacteria was dissolved in 500 μl of ultrapure water (from Roche Diagnostics). Take 8 μl of each bacterial dilution as template solution.
[0149] 2.3...
Embodiment 3
[0151] Example 3: Primers and assay design
[0152] 3.1 Screening of target genes
[0153] Table 3: Target genes used to design serotype-specific primer / probe pairs
[0154]
[0155] 3.2 Primer and probe sequence design
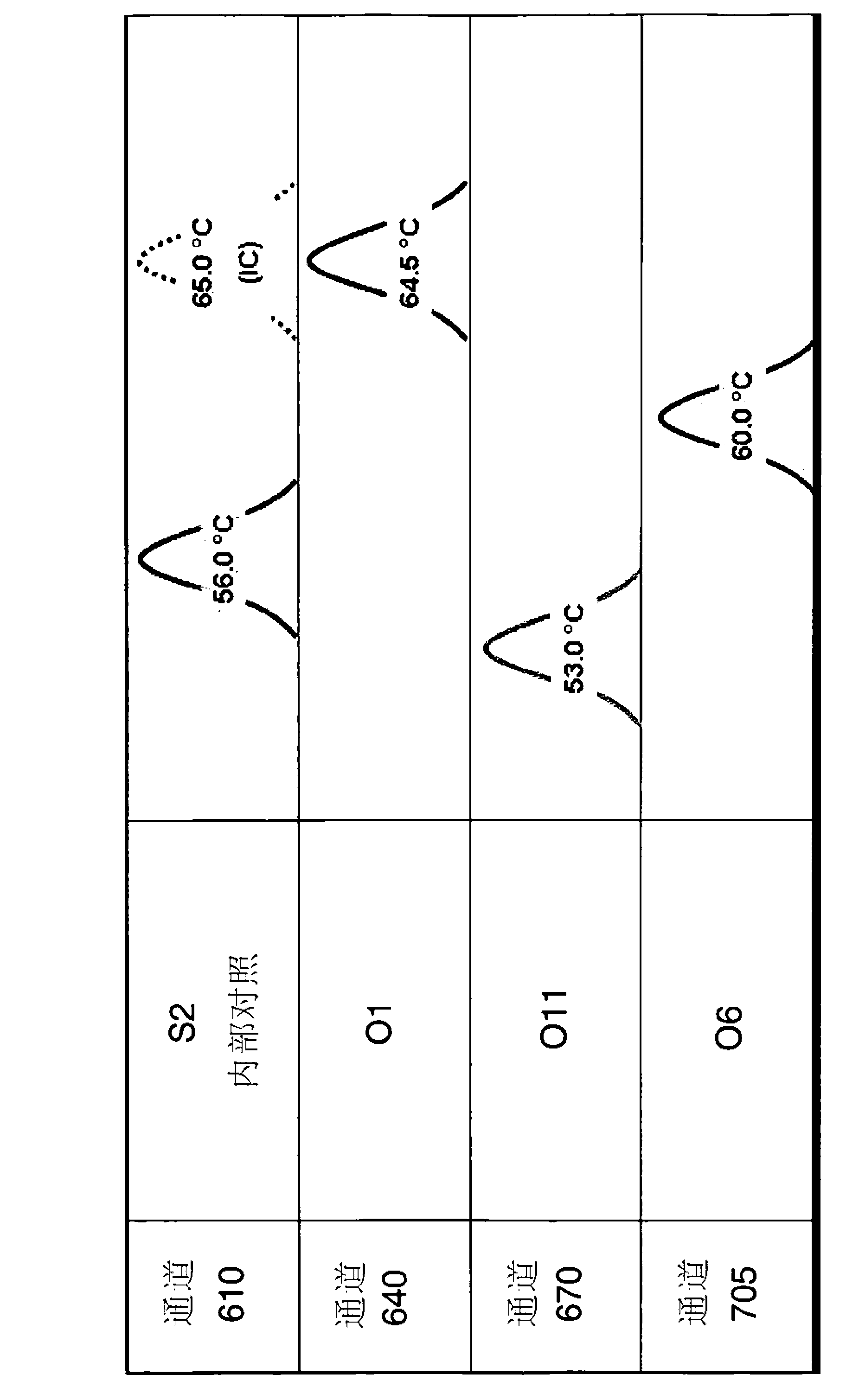
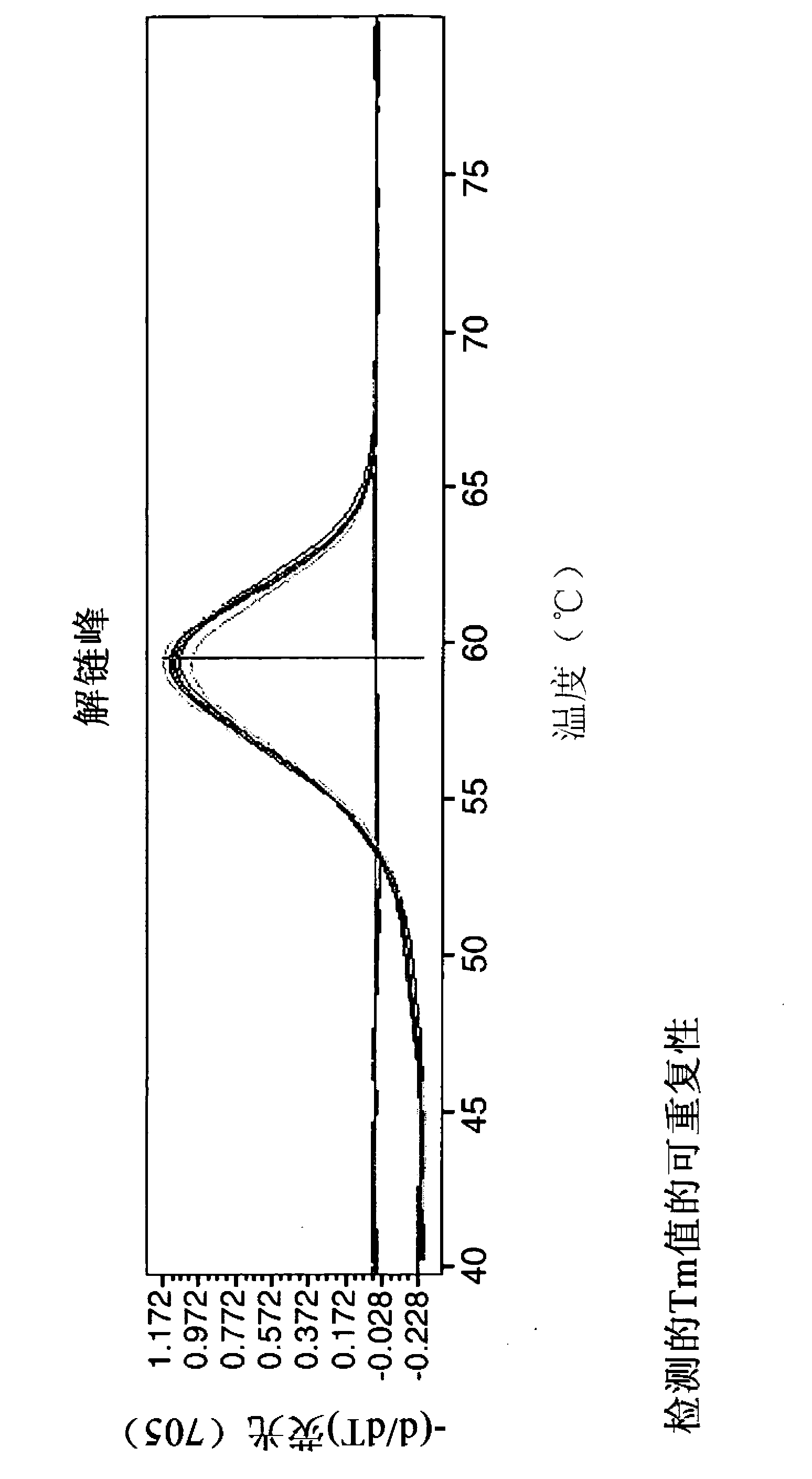
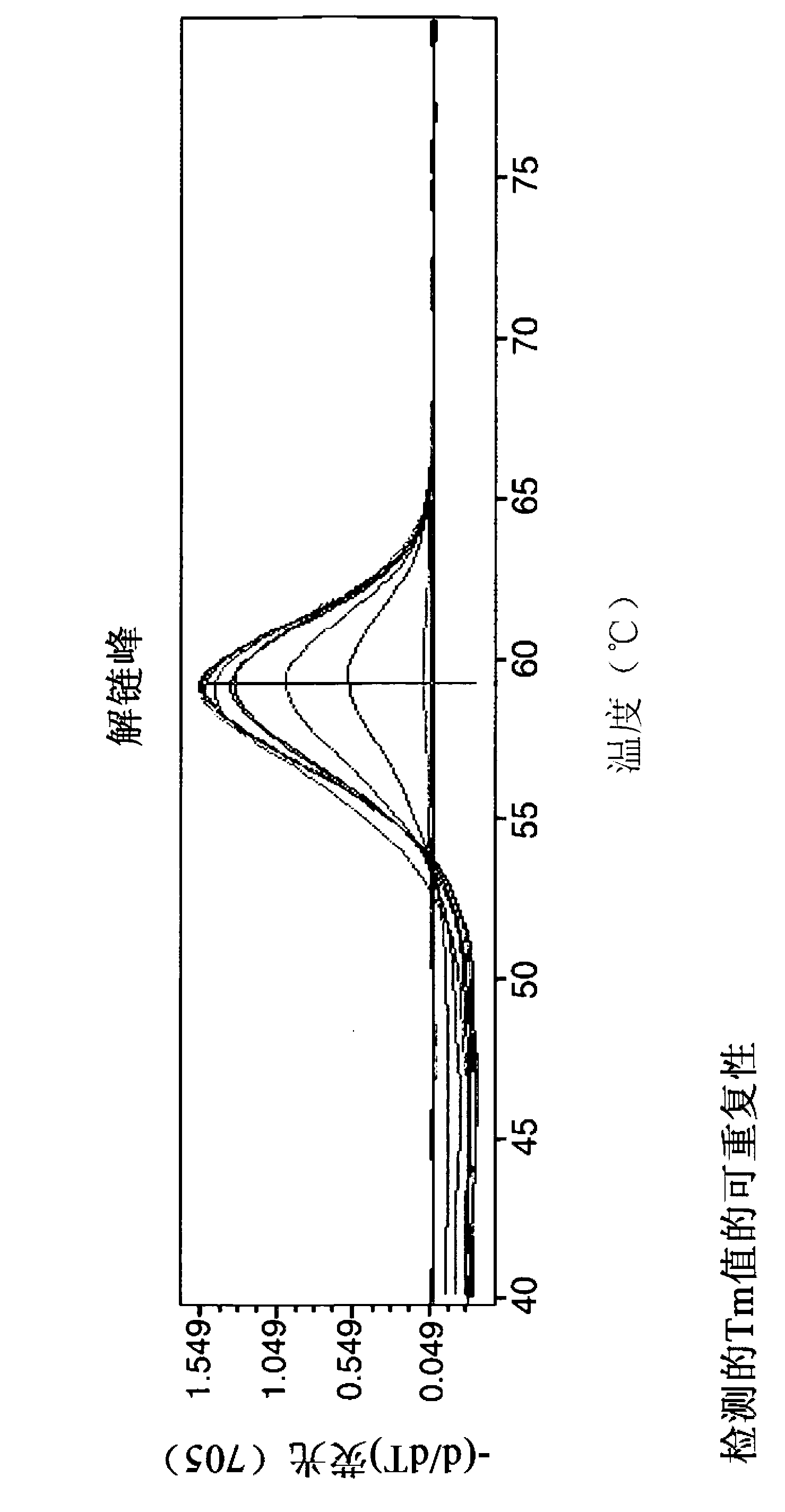
[0156] Primer / probe oligonucleotides were designed with the software LightCycler Probe Design Software 2.0 (from Roche Applied Science). Based on the software design, serogroup 2-specific probes were optimized by obtaining spacing between probe pairs, and serotype O11-specific probes were improved by shortening and perturbing sensing probes.
[0157] Table 4: Quadrivalent Pseudomonas aeruginosa serotyping assay - primer and probe sequences
[0158] In order to specifically detect one serotype in a test sample, two sets of oligonucleotide sequences are used: 2 primers and 2 probes. To specifically detect, for example, the 4 most prevalent Pseudomonas aeruginosa serotypes mentioned above, multiple primers and probes were used in one experiment.
[0159]...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com