Method for detecting CHO cell DNA
A cell and detection sensitivity technology, which is applied in the field of CHO cell DNA detection, can solve the problems of sample pretreatment, different primer and probe designs, and no unified standard substance, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0136] Embodiment 1. evaluates the performance of the primer pair shown in SEQ ID NO:3 and SEQ ID NO:4
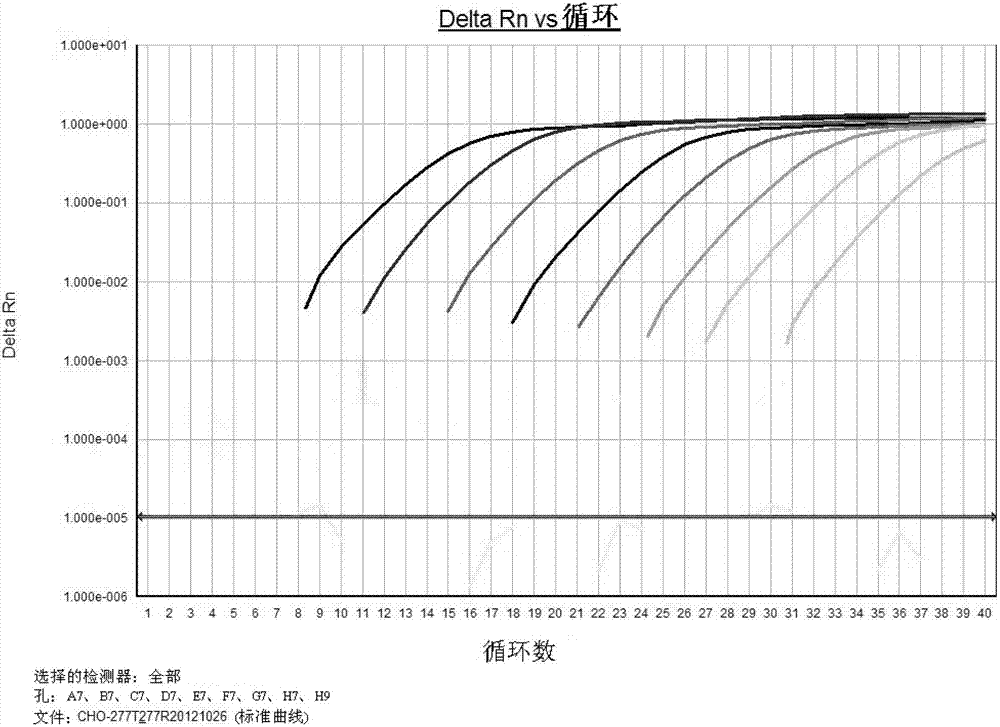
[0137] According to the above "Materials and methods used in the present invention", the primer pair shown in SEQ ID NO:3 and SEQ ID NO:4 was used for PCR detection. Experimental results such as figure 1 and figure 2 shown. in figure 1 is the reference product amplification curve, and the amplification curve shows an obvious exponential growth period. figure 2 is the reference standard curve. Depend on figure 2 It can be seen that when the concentration of the reference substance is 1000pg / μl, 100pg / μl, 10pg / μl, 1pg / μl, 100fg / μl, 10fg / μl, 1fg / μl and 0.1fg / μl, the slope of the standard curve drawn is -3.39 , the correlation coefficient (R2)=0.999, and the amplification efficiency was 97.23%. The standard curve has good linearity, and the detection sensitivity can reach 0.1fg / μl.
Embodiment 2
[0138] Example 2. Evaluation of the specificity of the primer pair shown in SEQ ID NO:3 and SEQ ID NO:4
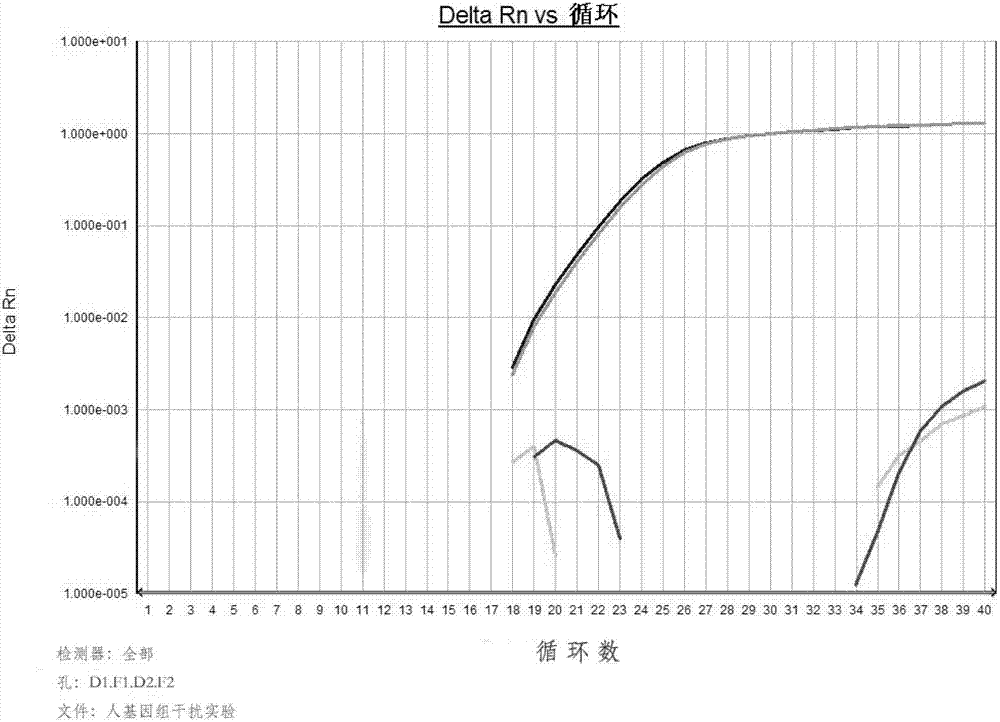
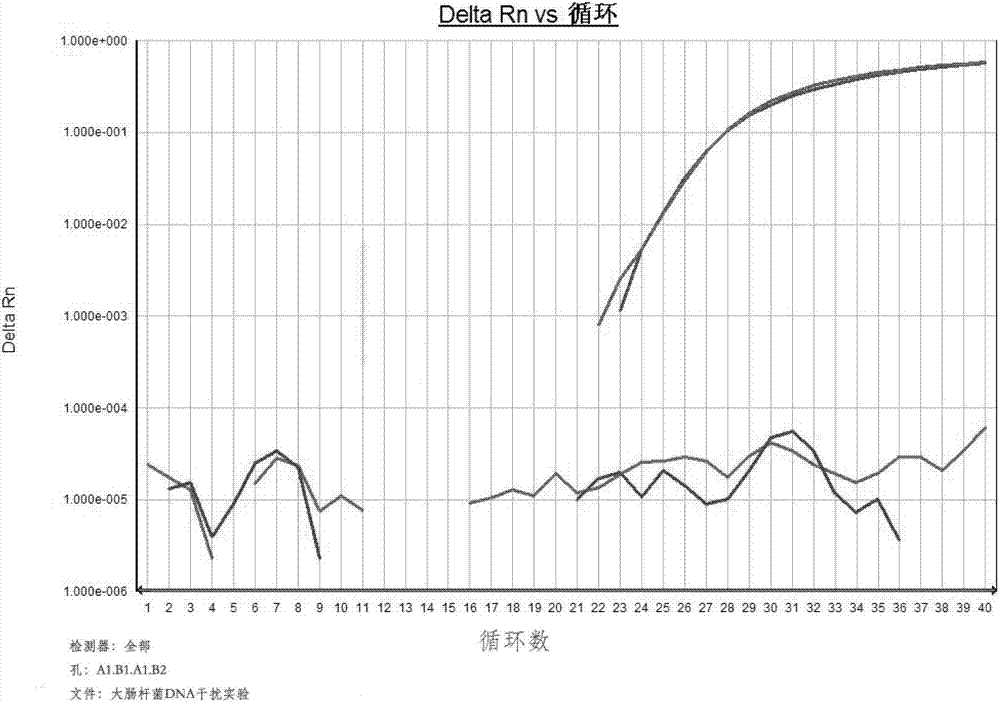
[0139] In order to evaluate the specificity of the primer pair shown in SEQ ID NO:3 and SEQ ID NO:4, human and Escherichia coli contamination DNA common in production and mouse and rat genes with high homology to hamster cells were selected as system interference experiment. Specifically:
[0140] 1. Human and Escherichia coli contamination DNA system interference experiment
[0141] Add the same concentration of 10pg / μl CHO gene and human gene (human liver cancer cell SK-HEP-1 comes from the cell bank of the Type Culture Preservation Committee of the Chinese Academy of Sciences; extract with takara gene extraction kit) and the same concentration of 10pg in the detection system / μl CHO gene and Escherichia coli gene (E.coil DH5α strain from the Institute of Microbiology, Chinese Academy of Sciences, No. 1.1595, extracted with takara gene extraction reagent) were used as ...
Embodiment 3
[0147] Example 3. Evaluation of the performance of the primer pair shown in SEQ ID NO:5 and SEQ ID NO:6
[0148] According to the method described in Example 1, the performance of the primer pair shown in SEQ ID NO:5 and SEQ ID NO:6 was evaluated.
[0149] Experimental results such as Figure 7 and Figure 8 shown. in Figure 7 is the reference product amplification curve, and the amplification curve shows an obvious exponential growth period. Figure 8 is the reference standard curve. Depend on Figure 8 It can be seen that when the concentration of the reference substance is 1000pg / μl, 100pg / μl, 10pg / μl, 1pg / μl, 0.1pg / μl and 0.01pg / μl, the slope of the standard curve drawn is -3.33, and the correlation coefficient (R2)= 0.997, the amplification efficiency was 99.66%. The standard curve has good linearity, and the detection sensitivity can reach 10fg / μl.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com