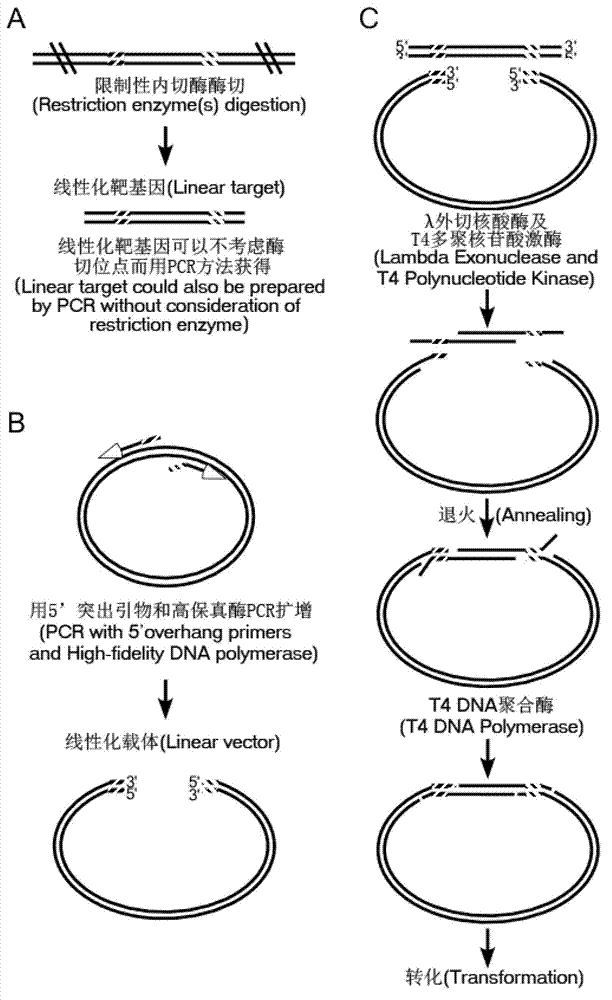
Method for simulating recombination and non-trace cloning
A cloning method and template technology, applied in the direction of recombinant DNA technology, the use of vectors to introduce foreign genetic material, etc., can solve the problems of high mutation rate, non-specific recombination of target genes, low amplification efficiency, etc., and achieve the effect of reducing mutations
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
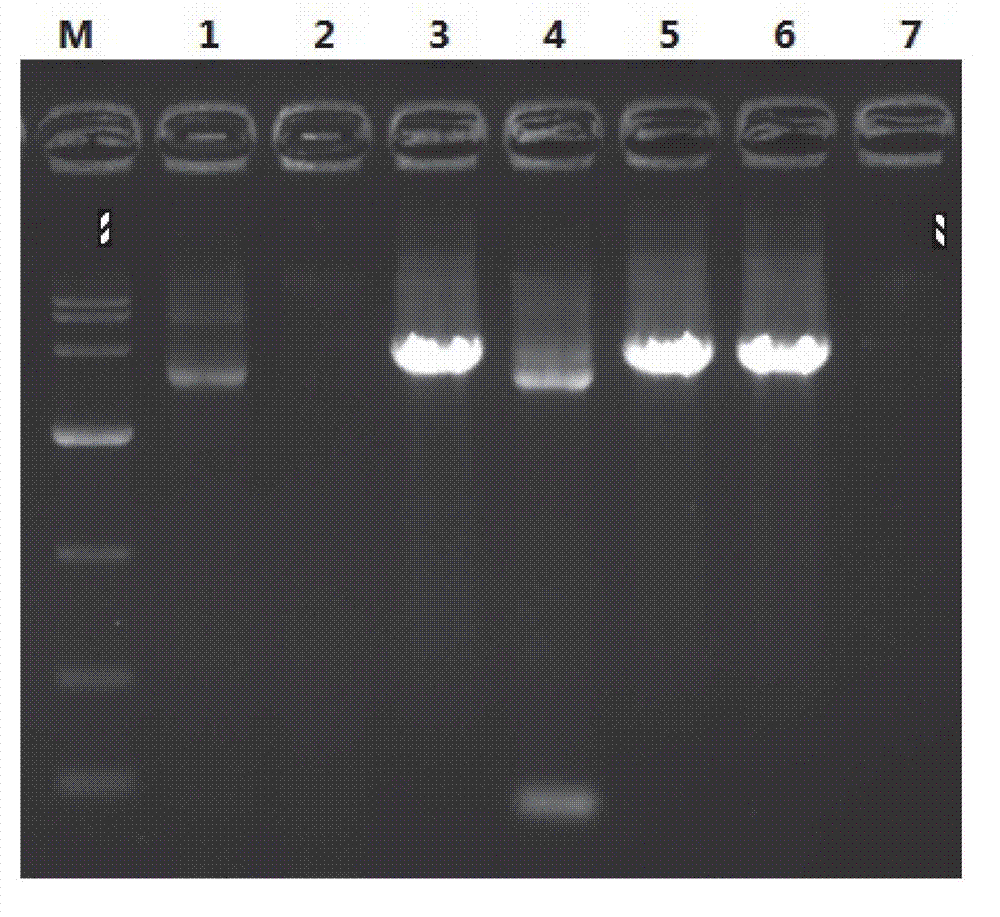
[0034] 1.1. Detect the efficiency of simulated recombinant traceless clones.
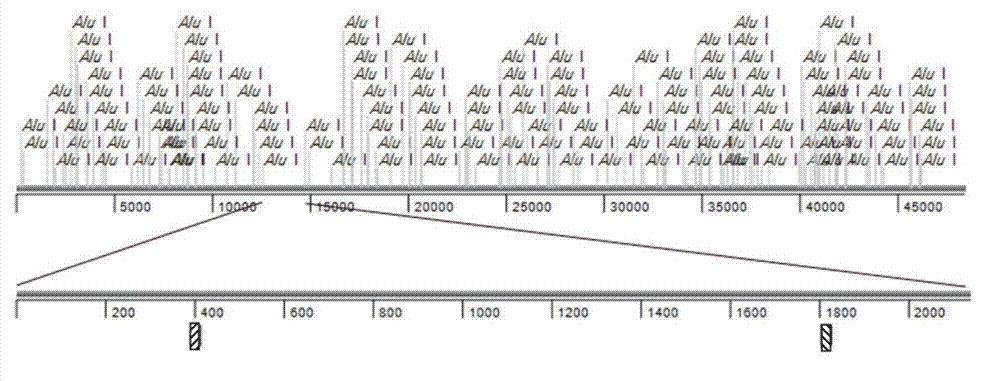
[0035] First, a segment ( figure 2 ) to test efficiency.
[0036] 1.1.1. Preparation of target DNA fragments.
[0037] In 40μl 1×NEB Buffer (New England Biolabs), 2μg Lambda DNA (Sangon) was incubated with 20U AluI endonuclease (New England Biolabs) at 37°C React for 4 hours to completely digest the Lambda DNA. Then heat to 70° C. and incubate for 10 minutes to inactivate the endonuclease, and the product obtained is a Lambda DNA digestion product with a final concentration of 50 ng / μl.
[0038] Table 1: Primers used to detect the efficiency of mock recombination scarless cloning
[0039] Overlap Primers Sequences (5'-3') 0 bp 0bpP1 ATCGAATTCCTGCAGCC 0bpP2 ATCAAGCTTATCGATACCG 15bp 15bpP1 TGCCGTCGTTGTTAA ATCGAATTCCTGCAGCC 15bpP2 AGAGTGATTTGCCGT ATCAAGCTTATCGATACCG 20bp 20bpP1 GTTCGTGCCGTCGTTGTTAA ATCGAATTCCTGCAGCC 20bpP...
Embodiment 2
[0068] 2.1. Clone the Sox10 ORF into pGEX-4T-1 by using the analog recombination traceless cloning method
[0069] The coding region (ORF) of the Sox10 gene has 1401bp, and the GC content reaches 63.24%. It failed to add a restriction site and its protective base at the 5' end of the conventional primer, and used the high-fidelity Phusion DNA polymerase (New England Biolabs) to amplify the target gene fragment by PCR, and then ligate it into the pGEX-4T-1 vector after enzyme digestion. Therefore, the Sox10 ORF was subcloned from the pBlueScript II KS(-) vector into the pGEX-4T-1 vector using the simulated recombination scarless cloning method to obtain the GST-Sox10 fusion expression plasmid.
[0070] 2.1.1. Preparation of target DNA fragments.
[0071] In 20 μl NEB buffer 4+BSA (New England Biolabs) reaction system, add 1 μg pBlueScript II KS(-) plasmid containing Sox10 ORF and use 10 U of endonucleases EcoRI-HF and XhoI (New England Biolabs) at 37°C React for 4 hours, th...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com