Method for determining type of microtubule-associated protein-1 light chain 3 protein spot
A technology for microtubule-related proteins and spots, which is used in material excitation analysis, fluorescence/phosphorescence, etc. to improve accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
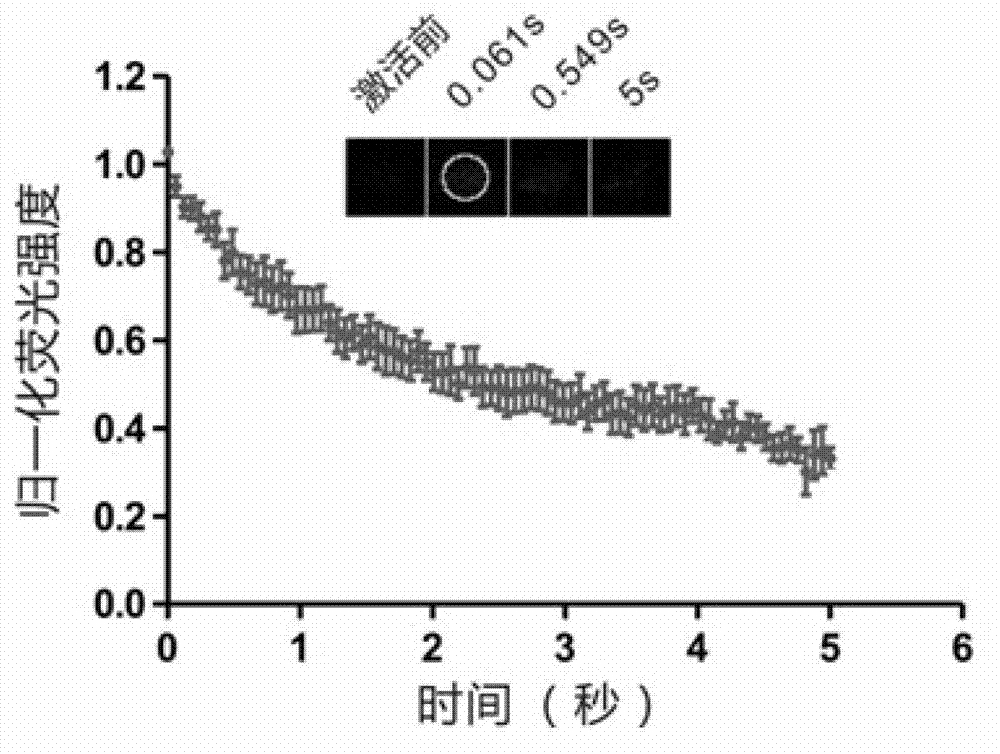
[0037] In this example, the LC3 protein (dendra2-LC3) fused to dendra2 photoconversion protein was first transfected into HeLa cells for 24 hours. Cells were then treated with puromycin for 2.5 hours, and dendra2-LC3 spots were induced. Puromycin is a widely used reagent to induce autophagic puncta formation. Finally, the FRAPa method was used for analysis. figure 1 It is the figure of FRAPa technology to determine the type of dendra2-LC3 spots induced by puromcyin. It can be seen from the figure that after the dendra2-LC3-labeled protein aggregates are activated by light, the fluorescence at the spots decreases rapidly and significantly.
Embodiment 2
[0039] In this example, GFP-fused LC3 protein (GFP-LC3) and mCherry-fused LC3 protein (mCherry-LC3G120A) were first co-transfected into HeLa cells for 24 hours. The cells were then treated with puromycin (Rapamycin) and chloroquine (CQ for short) for 6 hours. Rapamycin and CQ are widely used reagents to induce autophagic puncta formation. Finally, the FRAP method was used to analyze, figure 2 is the negative untreated group and the control group, image 3 are the graphs of CQ-induced GFP-LC3 / mCherry-LC3G120A puncta, Figure 4 Figures for rapamycin-induced GFP-LC3 / mCherry-LC3G120A puncta, Figure 5 is a graph of the statistics of the average intracellular GFP-LC3 / mCherry-LC3G120A spots in the negative control group, Image 6 is a graph of the statistics of the average intracellular GFP-LC3 / mCherry-LC3G120A spots in the CQ-treated group, Figure 7is a graph of the statistics of the average intracellular GFP-LC3 / mCherry-LC3G120A spots in the rapamycin-treated group, Figur...
Embodiment 3
[0041] In this example, GFP-LC3 and mCherry-LC3G120A were first co-transfected into HeLa cells for 24 hours. Cells were then treated with LY294002, wortmannin and MG132 for 6 hours. LY294002, wortmannin and MG132 are reported chemicals that induce protein aggregates. Finally, the FRAP method was used to analyze, Figure 10 Figures for LY294002-induced GFP-LC3 / mCherry-LC3G120A puncta, Figure 11 Figures for wortmannin-induced GFP-LC3 / mCherry-LC3G120A puncta, Figure 12 Figures for MG-132-induced GFP-LC3 / mCherry-LC3G120A puncta, Figure 13 Detection of LY294002-induced GFP-LC3 for FRAP technology + / mCherry-LC3G120A + Graph of the kinetics of spotted GFP-LC3; Figure 14 Detection of wortmannin-induced GFP-LC3 by FRAP technique + / mCherry-LC3G120A + Graph of the kinetics of spotted GFP-LC3; Figure 15 Detection of MG-132-induced GFP-LC3 for FRAP + / mCherry-LC3G120A + Graph of the kinetics of spotted GFP-LC3. from Figures 10 to 15 As can be seen, a certain amount of ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com