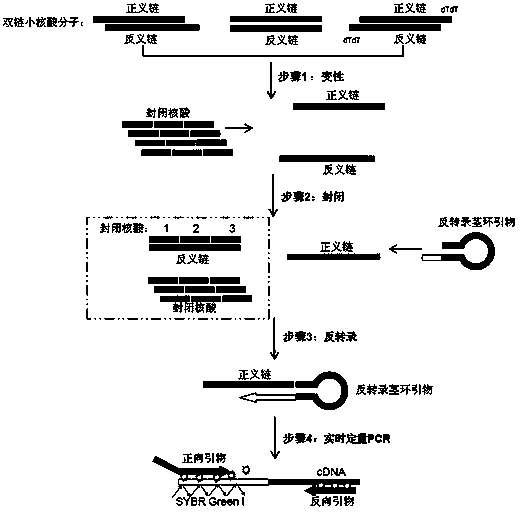
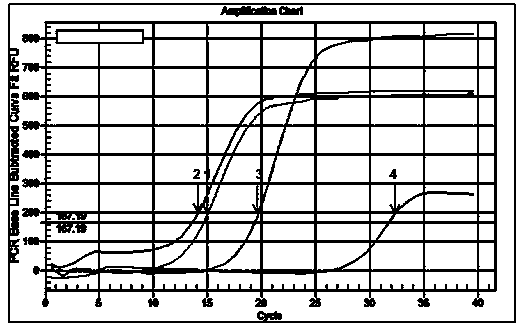
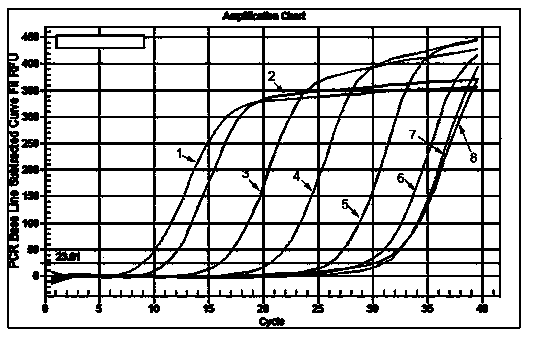
Small RNA detection method and use thereof
A detection method and small nucleic acid technology, applied in the field of small nucleic acid detection, can solve the problems of narrow detection range, high detection limit and high cost, and achieve the effects of low detection cost, high detection efficiency and strong specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment l
[0042] Identifying the Role of Blocking Nucleic Acids in Small Nucleic Acid Detection
[0043] 1 Main reagents, materials and instruments
[0044] 1.1 The si-NC sense strand and antisense strand (see Table 1 for the sequence) were synthesized by Biomaike Biotechnology Co., Ltd. Dilute the sense strand (single-strand RNA, ssRNA) and the annealed double strand (double-strand RNA, dsRNA) of si-NC to a concentration of 1 μM, respectively.
[0045] Table 1. si-NC sequences
[0046] 1.2
[0047] 1.3 The closed nucleic acid (DNA sequence) of si-NC: according to the 5'-3' direction, the closed nucleic acid complementary to the antisense strand of si-NC is designed, the length is 7nt, the sequence is shown in Table 2, provided by Biomaike Biotechnology Co., Ltd. synthesis.
[0048] Table 2. Blocking nucleic acids for si-NC
[0049] Blocked Nucleic Acid Name Sequence (5'-3') Length (nt) NC-B1 AATTCTC (SEQ ID NO: 3) 7 NC-B2 CGAACGT (SEQ ID NO: 4) 7 ...
Embodiment 2
[0071] Detection of double-stranded small nucleic acid (taking si-NC as an example)
[0072] 1 Main reagents, materials and instruments
[0073] 1.1 Sense and antisense strand sequences of si-NC (Table 1), synthesized by Biomaike Biotechnology Co., Ltd.
[0074] 1.2 The closed nucleic acid (DNA sequence) of si-NC: according to the 5'-3' direction, the closed nucleic acid complementary to the antisense strand of si-NC is designed. synthesis.
[0075] 1.3 Quantitative PCR detection primers were synthesized by Biomaike Biotechnology Co., Ltd.
[0076] Forward primer: 5'-TGCCCTTCTCCGAACGTGTC-3' (SEQ ID NO: 6);
[0077] Reverse primer: 5'-GTGCAGGGTCCGAGGT-3' (SEQ ID NO: 7);
[0078] Reverse transcription stem-loop primer: 5'-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAAACGTGA-3' (SEQ ID NO: 8).
[0079] 1.4 Reagents: EzOmics TM One-Step qPCR kit (Biomicro Biotechnology Co., Ltd.), etc.
[0080] 1.5 Instruments: real-time quantitative PCR instrument (Bio-Rad Company); centr...
Embodiment 3
[0097] Detection of small nucleic acids with different ends
[0098] 1 Main reagents, materials and instruments
[0099] 1.1 The 3' end of the si-NC sense strand is UU's si-NC-M1 (Table 5), which was synthesized by Biomaike Biotechnology Co., Ltd. The sense and antisense strands were annealed into double strands and diluted to a concentration of 10 μM.
[0100]Table 5. Modified si-NC sequences
[0101] 1.2
[0102] 1.3 The closed nucleic acid (DNA sequence) of si-NC: according to the 5'-3' direction, the closed nucleic acid complementary to the antisense strand of si-NC is designed, the length is 7nt, the sequence is shown in Table 2, provided by Biomaike Biotechnology Co., Ltd. synthesis.
[0103] 1.4 Quantitative PCR detection primers were synthesized by Biomaike Biotechnology Co., Ltd.
[0104] Forward primer: 5'-TGCCCTTCTCCGAACGTGTC-3' (SEQ ID NO: 6);
[0105] Reverse primer: 5'-GTGCAGGGTCCGAGGT-3' (SEQ ID NO: 7);
[0106] Reverse transcription stem-loop primer: 5...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com