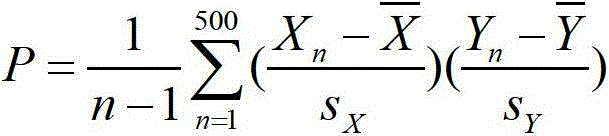Method for detecting medicine adverse reaction with genome expression profiling
A technology of genome expression and adverse reactions, applied in special data processing applications, instruments, electrical digital data processing, etc., can solve problems such as hysteresis and achieve good application prospects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
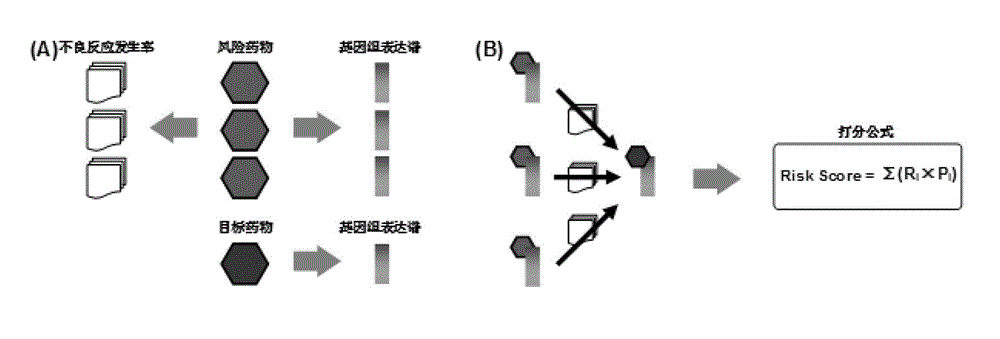
[0035] Establishment of pharmacogenomic expression profiling and selection of risky drugs
[0036] Human tumor cell lines (including MCF7, PC3, HL60, etc.) were treated with each of 1309 drugs for comparison with untreated cells. The ribonucleic acid (abbreviated as RNA) in the cell is separated and used to synthesize complementary deoxyribose nucleic acid (cDNA). Through the Affymetrix HG-U133A gene chip, the cDNA was hybridized with the probe to determine the abundance of the RNA corresponding to each cDNA. By comparing the RNA abundance levels of drug-treated cells and untreated cells, a total of 22,283 RNAs can be determined as a result of drug treatment. ConnectivityMap database).
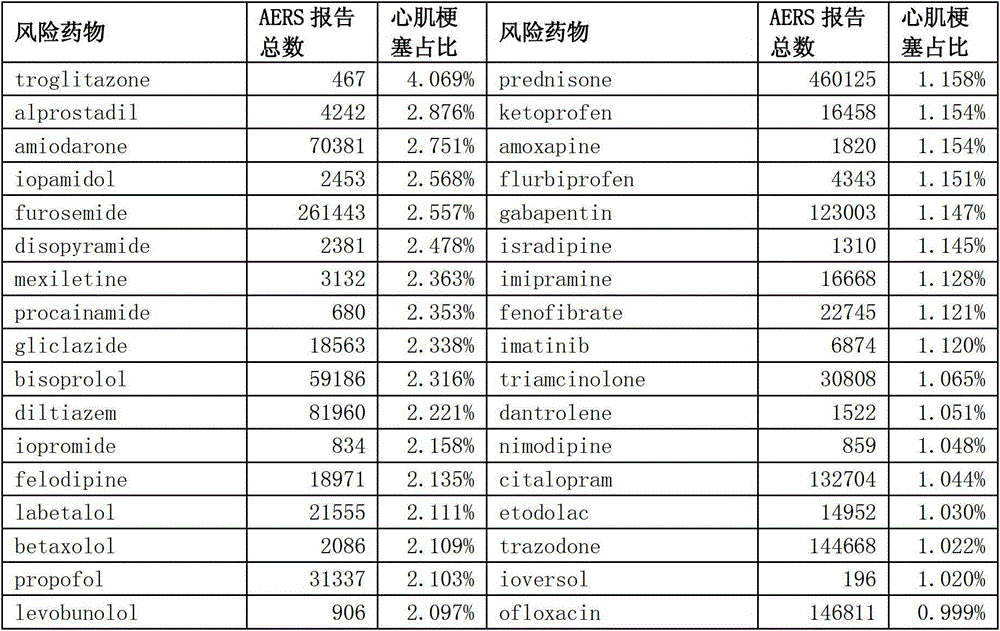
[0037] Among the 1309 treated drugs, 128 risk drugs tend to cause myocardial infarction (Myocardial Infarction), such as Troglitazone, Atropine, and Imatinib, were obtained by consulting the drug instructions (http: / / sideeffects.embl.de / ). By querying the Adverse Event Reporting System (AER...
Embodiment 2
[0042] Predicting the risk of adverse reactions of target drugs based on the genomic expression profiles of risky drugs
[0043]Except for 128 risk drugs, the remaining 1181 drugs were used as test drugs. Based on the similarity between the test drug and the risk drug in the genome expression profile (such as Pearson correlation coefficient, etc.), and weighting the proportion of myocardial infarction adverse reaction reports for each risk drug of similarity, the myocardial infarction risk of the test drug can be calculated coefficient. Its formula is as follows:
[0044] Score = Σ i = 1 128 R i × P i
[0045] R in the above formula i Represents the proportion of myocardial infarction adverse reaction reports of the i-th risk drug, P i Represents the Pearson correlation coef...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com