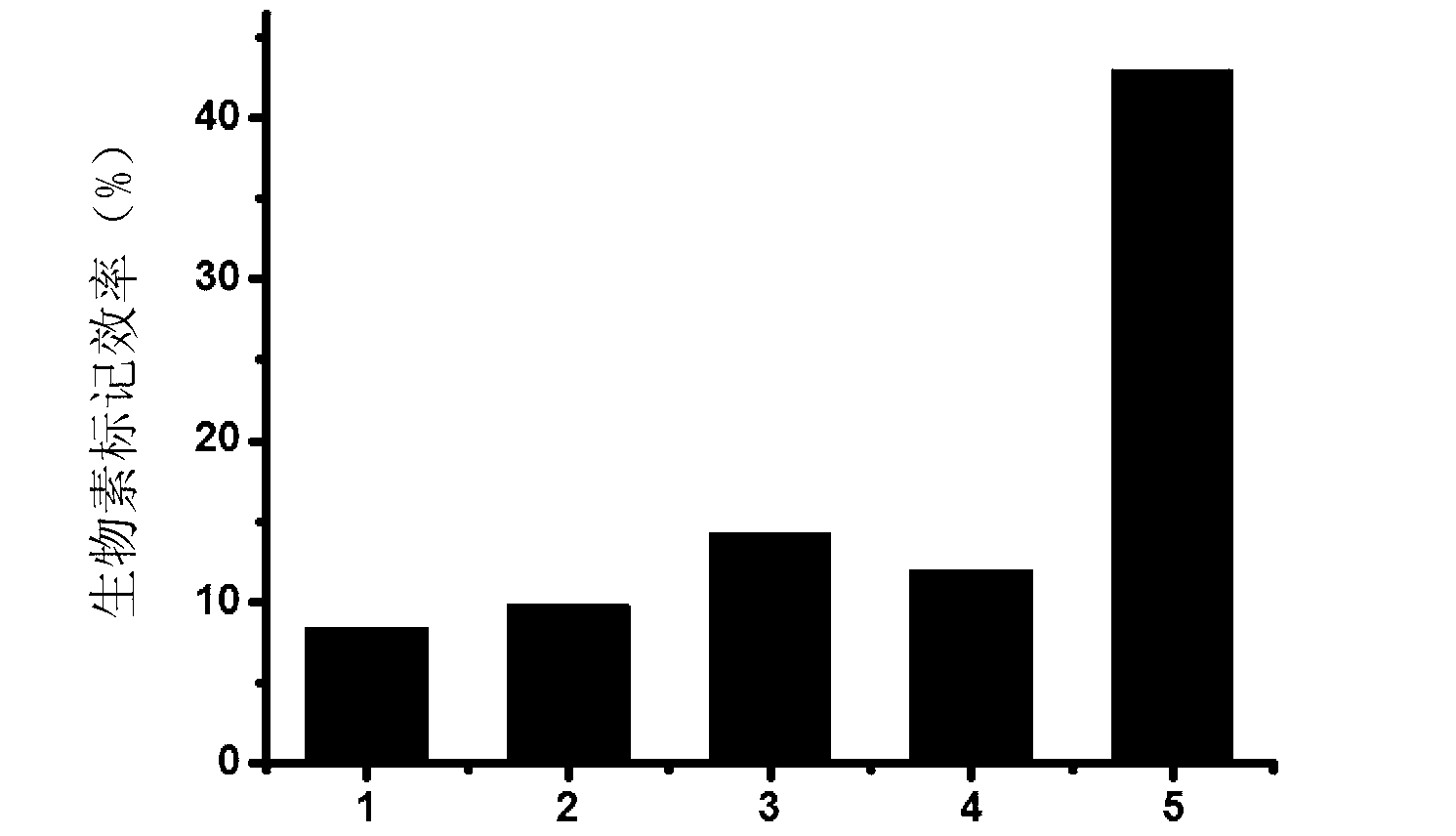
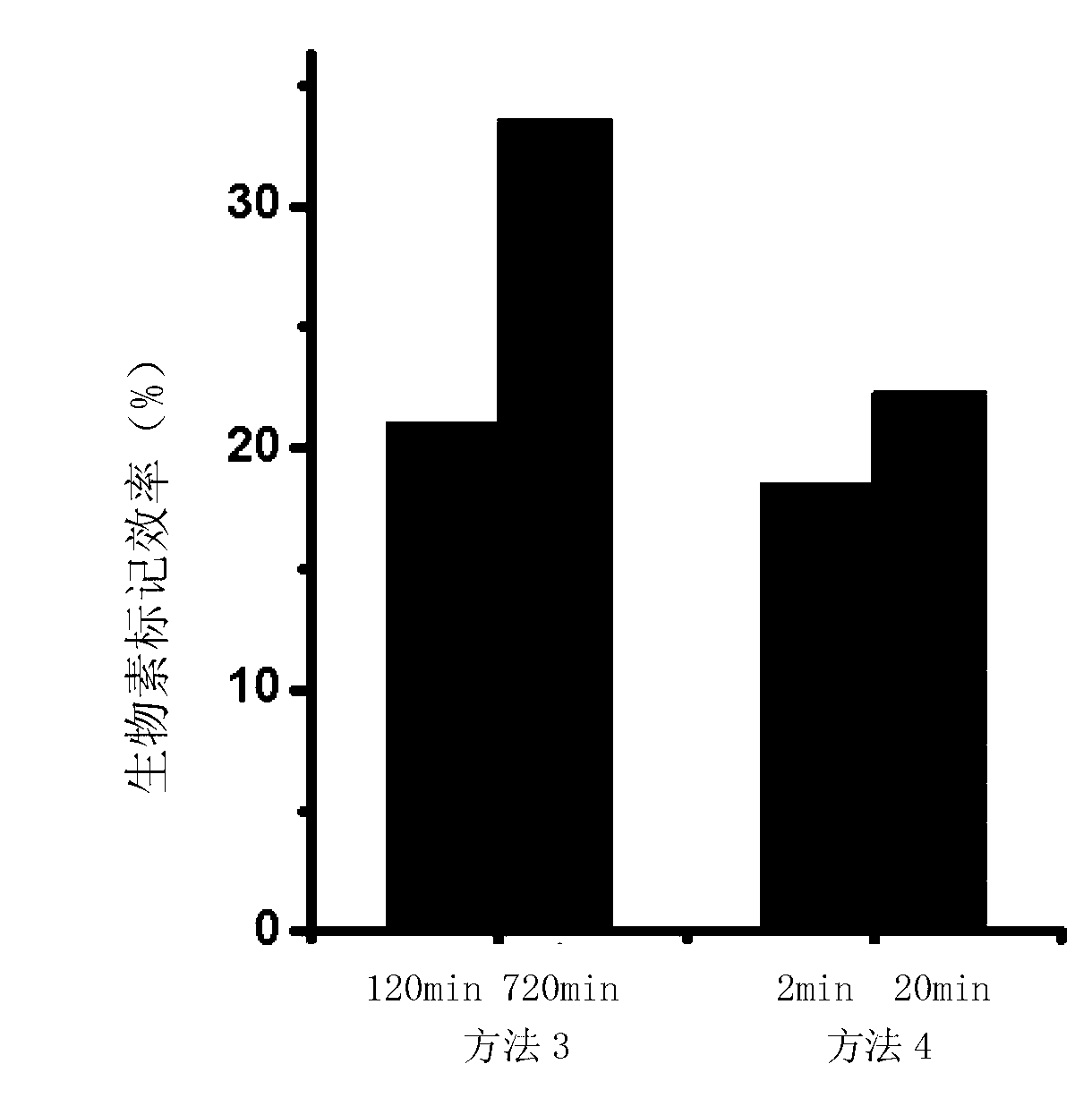
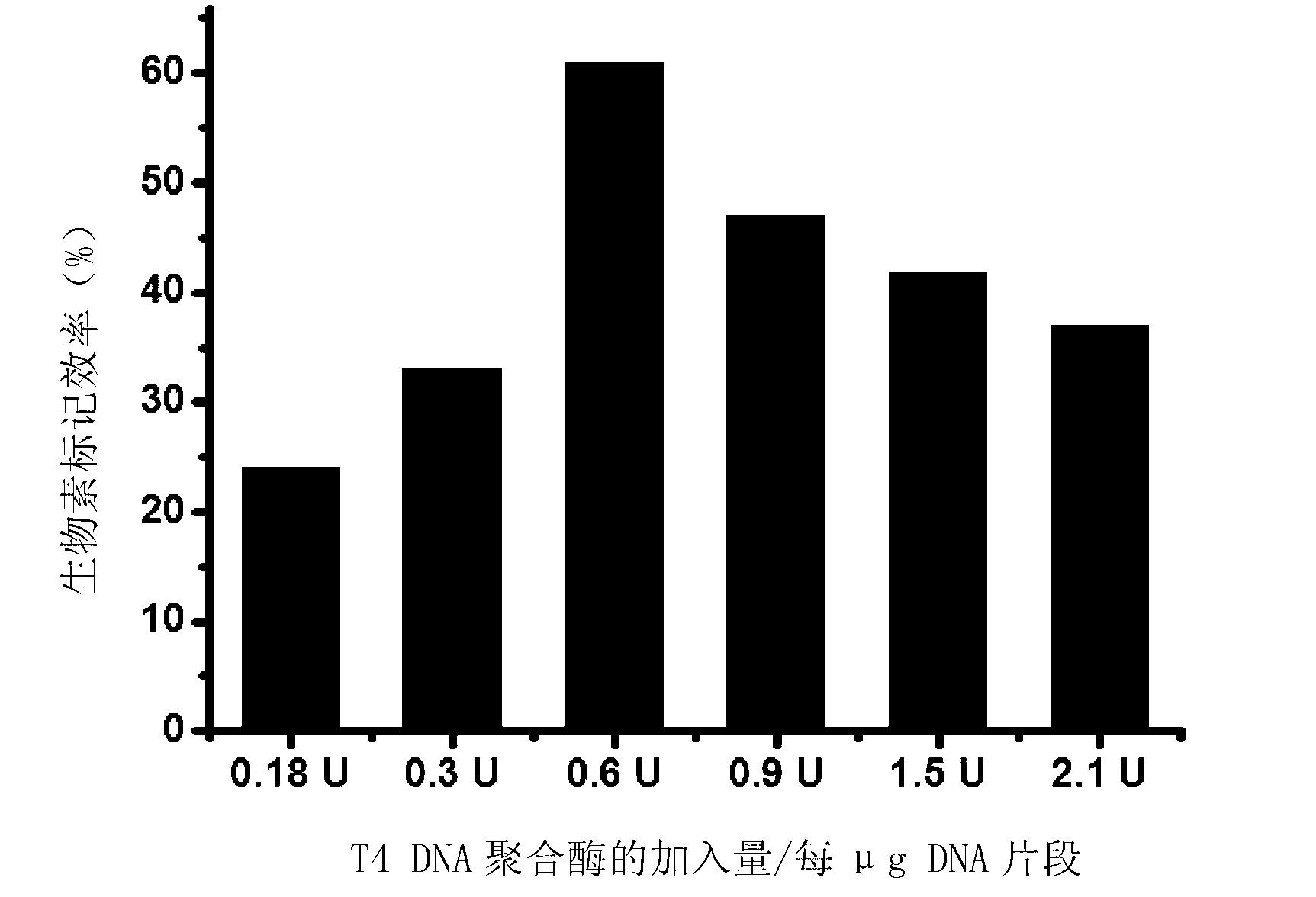Method for performing 3' end biotin labeling on DNA fragments
A technology of biotin labeling and fragmentation, applied in the direction of DNA preparation, recombinant DNA technology, etc., can solve the problems of low labeling efficiency and achieve the effects of wide application range, reduced sample loss and preference, and high labeling efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
[0058] Preparation method of PBS buffer solution: dissolve NaCl8g, KCl0.2g, KH in 800ml distilled water 2 PO 4 0.24g, Na 2 HPO 4 12H 2 O3.63g, adjust the pH value to 7.4 with HCl, add water to 1L, 1.034×10 5 Pa high-pressure steam sterilization for 20min, stored at room temperature.
[0059] The cell lysate (pH8.0) is composed of solvent and solute, the solvent is water, the solute and its concentration are: 10mM Tris-HCl, 10mM NaCl, 0.2% (volume ratio) Triton X-100 and 1× protease inhibitor.
[0060] The cell nucleus lysate (pH8.0) is composed of solvent and solute, the solvent is water, the solute and its concentration are: 50mM Tris-HCl, 10mM EDTA, 1g / 100mL SDS and 1× protease inhibitor.
[0061] 2×BW buffer (pH8.0) is composed of solvent and solute, the solvent is water, the solute and its concentration are: 10mM Tris-HCl, 1.0mM EDTA and 2.0M NaCl.
[0062] TWB buffer is composed of solvent and solute, the solvent is water, the solute and its concentration are: 5mM T...
Embodiment 1
[0063] Embodiment 1, preparation the DNA fragment that ultrasonic breaks up
[0064] 1. Formaldehyde cross-linked chromatin and harvested nuclei
[0065] 1. In order to imitate the DNA fragments produced by chromatin conformation capture technology as much as possible, MCF-7 cells were fixed with 1% formaldehyde and reacted at room temperature for 10 minutes.
[0066] 2. Discard the supernatant, add 10ml of PBS buffer solution containing 0.125M glycine, and react at room temperature for 10 minutes.
[0067] 3. Discard the supernatant, add 1ml 0.25% trypsin solution, incubate at 37°C for 10min, then add 0.4ml serum and 10μl protease inhibitor to stop the reaction.
[0068] 4. Scrape the cells and wash with PBS buffer.
[0069] 5. Add 40ml cell lysate and 100μl protease inhibitor, mix thoroughly, and incubate at 4°C for 10-30min.
[0070] 6. Centrifuge at 4°C and 1500g for 5 minutes to collect the precipitate, which is the nucleus with chromatin cross-linked and fixed by form...
Embodiment 2
[0076] Embodiment 2, the DNA fragment of preparation enzyme cut
[0077] 1. Extract the genomic DNA of MCF-7 cells,
[0078] 2. Digest the genomic DNA obtained in step 1 with restriction endonucleases, and recover DNA fragments of 100-1500 bp.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com