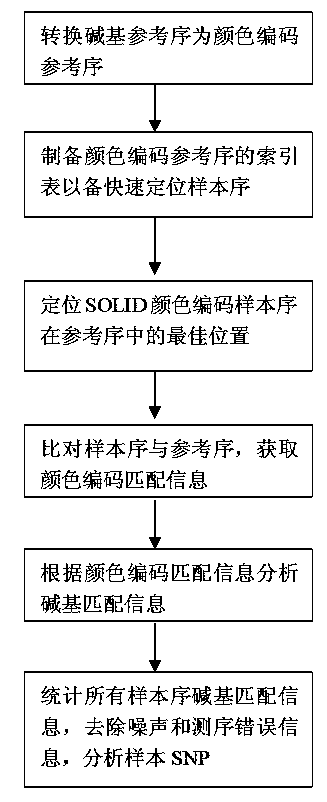
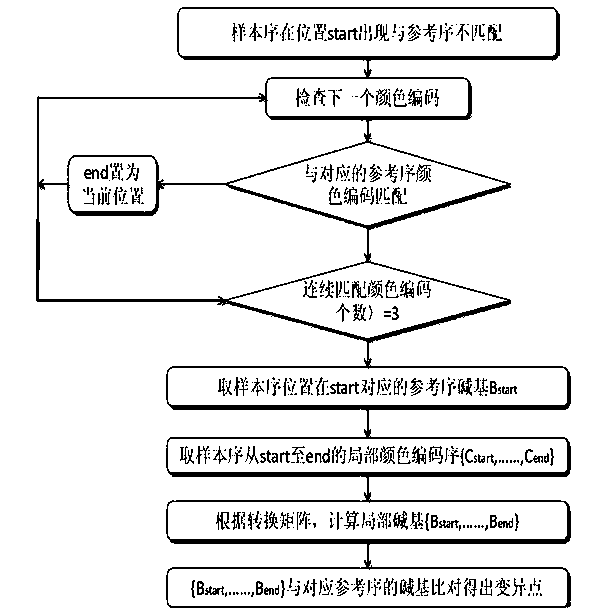
Gene SNP (single nucleotide polymorphism) site detection method based on SOLID (supported oligo ligation detection) sequencing technique
A detection method and site technology, applied in the field of bioengineering, can solve the problems of SNP site detection methods are difficult to meet, high-throughput sequencing technology has a large number, etc., to avoid false positive SNP sites, high anti-noise characteristics, and improve accuracy degree of effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
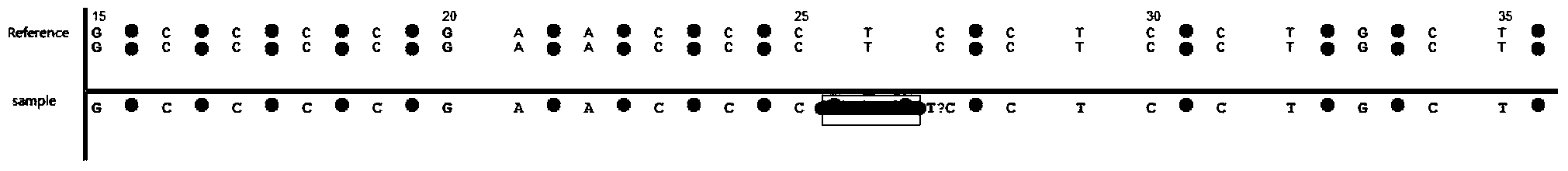
[0054] The HLA gene is the human leukocyte antigen gene, which is a highly polymorphic complex composed of a series of closely linked gene loci. In order to make the purpose, technical scheme and advantages of the present invention clearer, this paper takes the HLA type I gene HLA-A gene used for bone marrow transplantation matching as an example to perform data analysis:
[0055] 1. The data is downloaded from EBI (http: / / www.ebi.ac.uk / ipd / imgt / hla / dictionary.html). Including GeneBank format data of HLA-A (HLA-A.gbk), and fasta sequence data of each subtype of HLA-A
[0056] (HLA-A_dictionary.fasta).
[0057] 2. Extract the subtype of HLA-A 01:01:01:01 as the standard sequence (HLA-A_01010101.fasta).
[0058] 3. Comparing the sequence with the HLA-A.gbk reference sequence, a total of 45 SNP sites were identified, which were recorded as positive SNP sites
[0059] 4. Perform data simulation on the HLA-A 01:01:01:01 sequence, add 5% noise points randomly, and construct 2824 ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com