Non-invasive fetal genetic screening by sequencing analysis
A fetal and sequencing technology, applied in the field of non-invasive technology, can solve the problems of sample consumption, differentiation, and digital analysis that do not allow the detection of single nucleotide polymorphisms in fetuses, so as to achieve improved clinical performance and high detection rate Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
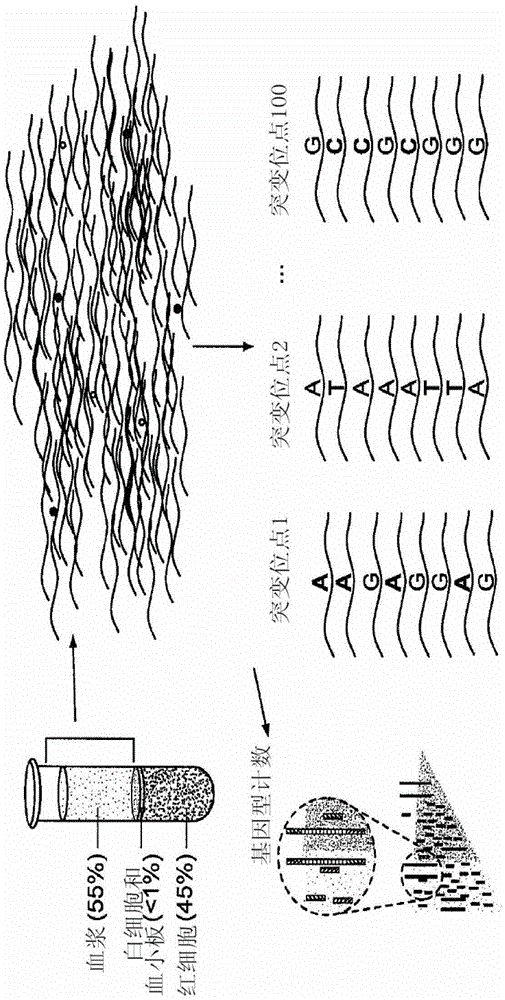
[0035] A. Biological sample preparation
[0036] The methods of the invention involve non-invasive detection. In a specific embodiment of the method, the starting material is maternal blood. In order to obtain sufficient DNA for testing, preferably 10-20 mL of blood is drawn to obtain approximately at least 10,000 genome equivalents of total DNA. This sample size was estimated based on fetal DNA present at approximately 25 genome equivalents / mL of maternal plasma in early pregnancy, a concentration of fetal DNA of approximately 3.4% of total plasma DNA. However, for genetic screens requiring less statistical significance, or DNA samples in which fetal DNA is enriched, less blood may be drawn.
[0037] It should be noted that although fetal DNA is referred to throughout this specification, fetal RNA found in maternal blood can also be analyzed. hPL (human placental lactogen) and hCG (human chorionic gonadotropin) mRNA transcripts, as analyzed using the respective real-time ...
Embodiment 1
[0093] Example 1: Detection of fetal genetic abnormalities in maternal blood
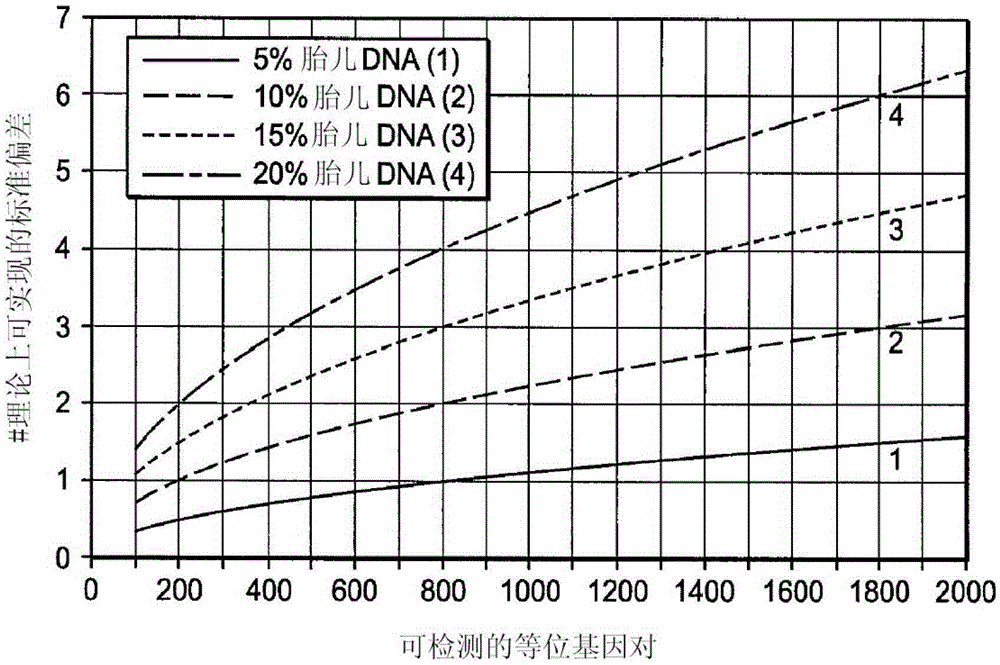
[0094] The amount of fetal DNA in maternal blood varies widely and increases as pregnancy progresses. In early pregnancy, the levels of fetal DNA present in maternal blood are insufficient to discern differences in the ratios of maternal and fetal DNA. The difference in the proportion of maternal and fetal DNA in maternal blood can be estimated.
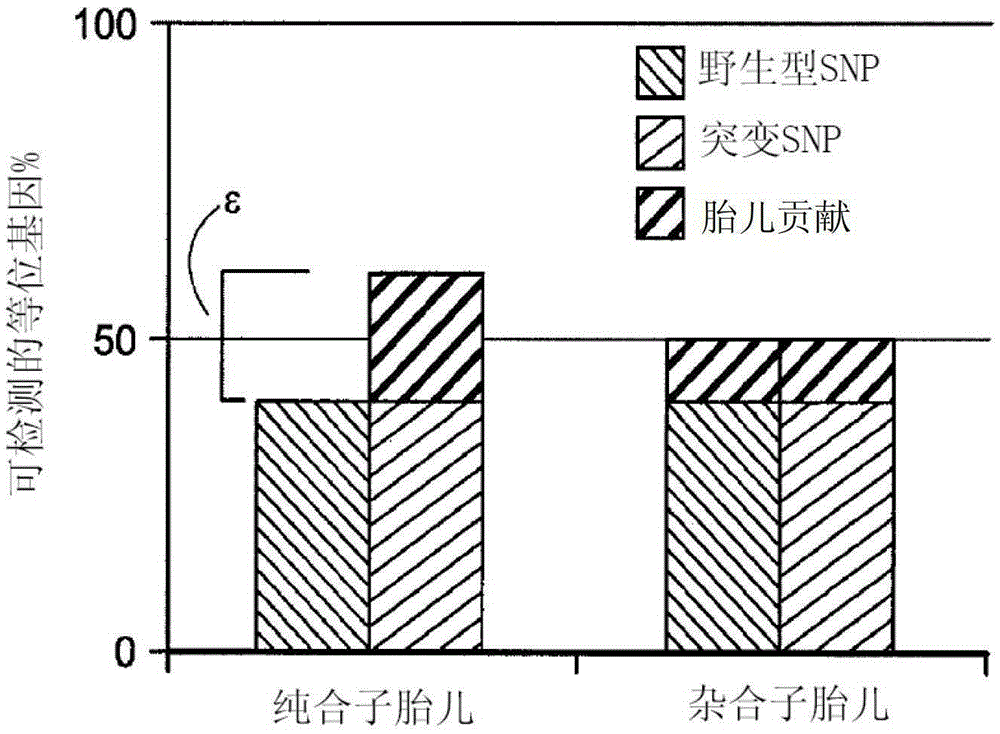
[0095] The finite target allele counts according to the Poisson distribution are calculated as follows:
[0096] Interpretation:
[0097] N T =#total alleles=N M +N W ,
[0098] N M = # mutant allele, N W =# wild type allele
[0099] The above formula is shown in figure 2 , which shows the theoretical fetal count mean, assuming maternal carrier of the mutation, ε is the fraction of detectable alleles equal to the difference between wild-type and mutant alleles in homozygous fetuses.
[0100] Homozygous case: N M –N W =ε*N T
[0101] Cas...
Embodiment 2
[0114] Example 2: Avoiding amplification bias when screening a panel of genetic mutations
[0115]Amplification bias is a known problem in multiplex PCR. Amplification bias can be addressed by referring to one genotype within the amplicon relative to another. However, in order to obtain the fraction of fetal DNA in a sample, comparisons between different amplicons are necessary, each of which will experience amplification bias (e.g., the amount of two different targets that are quantitatively different before amplification equal, 2-fold or higher difference after amplification).
[0116] Preliminary evidence suggests that digital PCR produces less amplification bias than Poisson sampling error for single amplicons. If a multiplex PCR reaction is to be spiked with a known or equal number of reference targets with known sequences, the reference targets can then be used to detect relative or absolute amounts of targets in different samples.
[0117] For example, if a multipl...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com