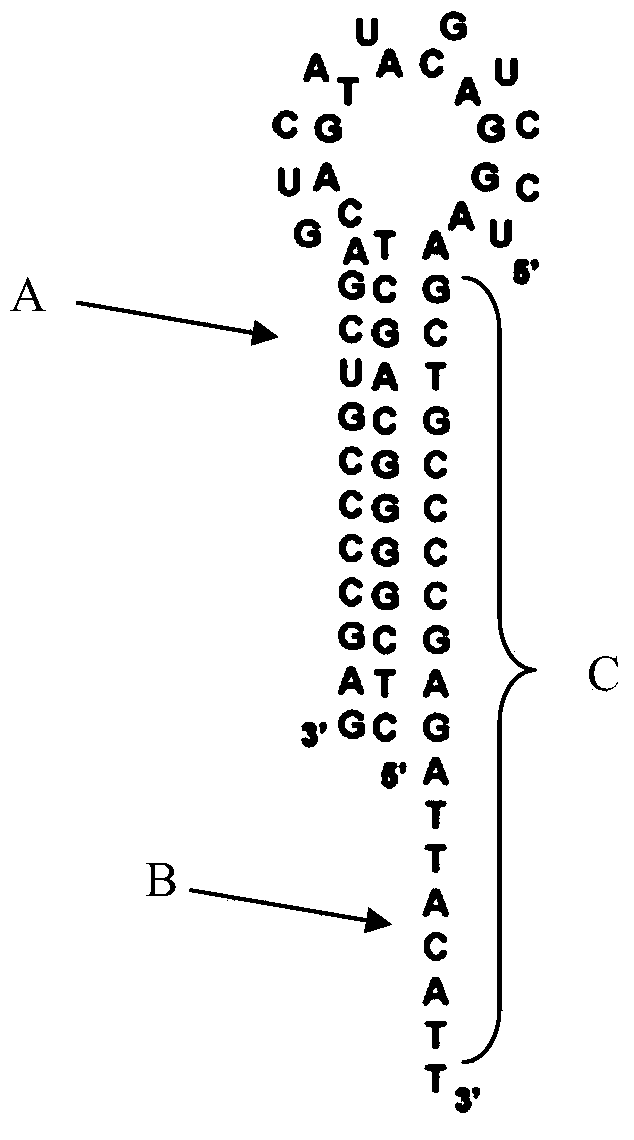
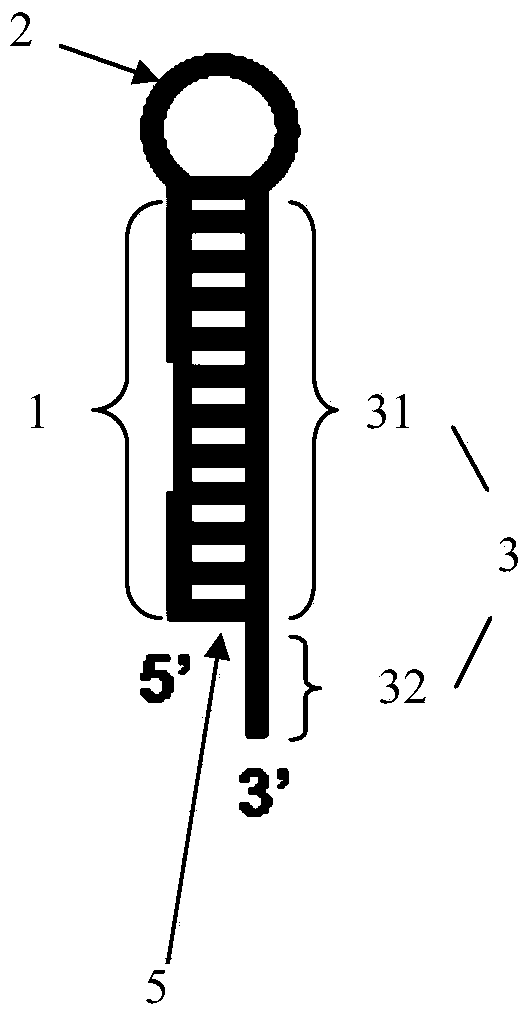
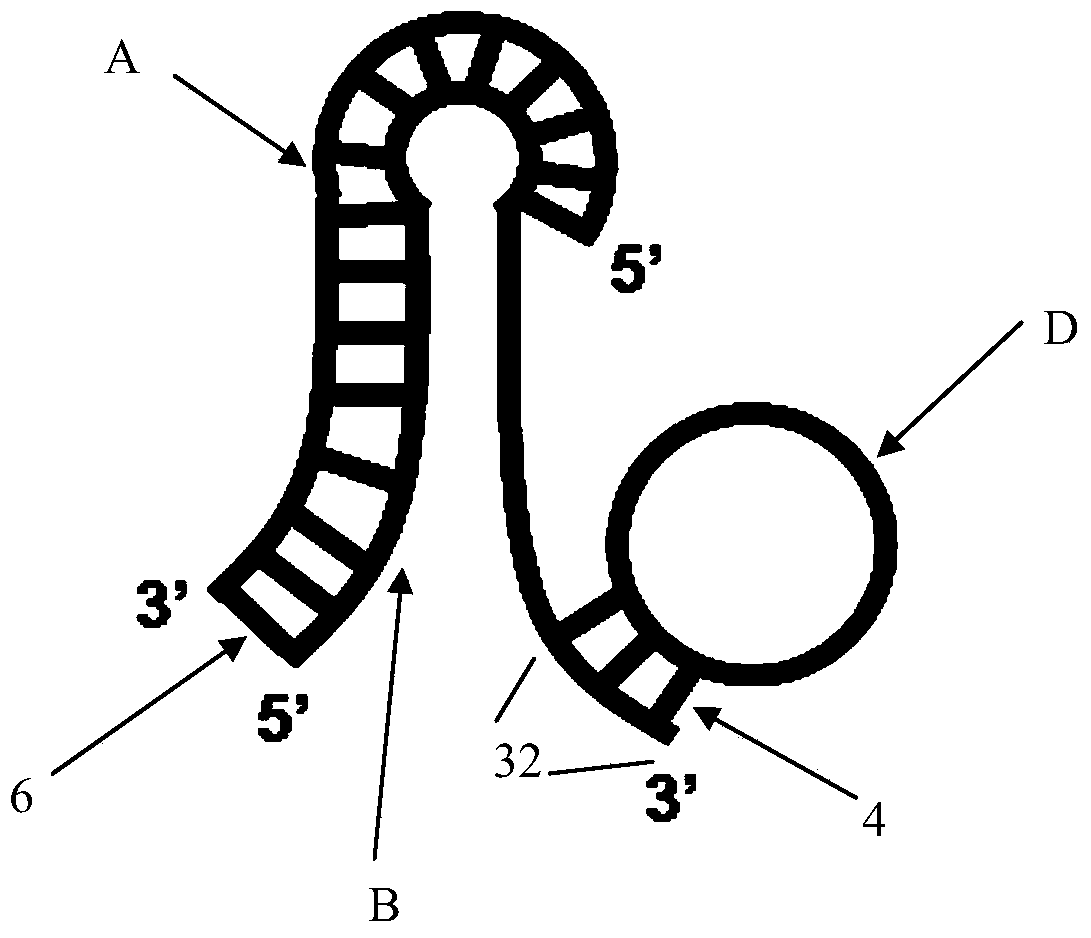
Rolling circle amplification based miRNA (micro ribonucleic acid) assay probe, assay method and kit
A rolling circle amplification reaction and detection probe technology, applied in the field of miRNA detection probes, can solve problems affecting detection specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0120] Provide hairpin probes, rolling circle probes, miR-4529-3p similar in sequence to miR-486-5p to be tested, and miR-486-5p precursors based on rolling circle amplification reactions for detecting miR-486-5p, including the following step:
[0121] (1) In vitro synthesis of miR-486-5p to be tested, miR-4529-3p similar in sequence to miR-486-5p to be tested, and miR-486-5p precursor; said miR-486-5p, miR-4529 The nucleotide sequences (5' to 3') of -3p and miR-486-5p precursors are shown in SEQ ID NO:3, SEQ ID NO:4 and SEQ ID NO:5, respectively;
[0122] (2) Design according to the sequence of miR-486-5p to be tested, and synthesize hairpin probes and circular probes in vitro; the nucleotide sequences (5' to 3') of the hairpin probes and circular probes are respectively As shown in SEQ ID NO:1 and SEQ ID NO:2, the nucleotide sequences (5' end to 3') of the miR-486-5p, miR-4529-3p and miR-486-5p precursor are respectively As shown in SEQ ID NO:3, SEQ ID NO:4 and SEQ ID NO:5...
Embodiment 2
[0132] Figure 5 The flow chart of detecting miR-486-5p using rolling circle amplification reaction provided for the embodiment of the present invention, combined with Figure 5 , the present embodiment provides a method for detecting miR-486-5p using rolling circle amplification reaction, comprising the following steps:
[0133] (1) Sample preparation
[0134] Take the miR-486-5p synthesized in Example 1 as the sample to be tested;
[0135] (2) Rolling circle amplification reaction
[0136] (2a) Provide or prepare experimental reagents:
[0137] 10U / ul Phi29 DNA polymerase, 40U / ul ribonuclease (RNase) inhibitor, 10mmol / ul dNTPs, 50×SYBR Green II dye, 10×Phi29 DNA polymerase reaction buffer (500mmol / L Tris -hydrochloric acid pH7.5, 100mmol / L magnesium chloride, 100mmol / L ammonium sulfate and 40mmol / L dithiothreitol), phosphate buffer (137mmol / L sodium chloride, 2.7mmol / L potassium chloride, 4.3mmol / L L disodium hydrogen phosphate and 1.4mmol / L potassium dihydrogen phospha...
Embodiment 3
[0163] The effect of rolling circle amplification reaction at different temperatures on the detection sensitivity of miR-486-5p, miR-4529-3p and miR-486-5p precursors includes the following steps:
[0164] (1) Sample preparation
[0165] Take miR-486-5p, miR-4529-3p and miR-486-5p precursor synthesized in Example 1 as samples to be tested;
[0166] (2) Rolling circle amplification reaction
[0167] (2a) Provide or prepare experimental reagents: refer to step (2a) of Example 2
[0168] (2b) Formation of the initial state card-issuing probe
[0169] Take the hairpin probe synthesized in vitro in Example 1, dilute it to 50nmol / L with 1× hairpin probe annealing buffer, incubate at 100°C for 8 minutes, and then slowly cool to room temperature (cooling for 4 hours) to make it fully folded and formed The card-issuing structure, the annealed card-issuing probe is the initial state of the card-issuing probe, and has a card-issuing structure;
[0170] (2c) Perform rolling circle amp...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com