Method for preventing and treating tumors by using microRNA sponge technology
A sponge and tumor technology, applied in the field of biomedicine, can solve problems such as the connection between NF-κB and cancer and the unclear mechanism
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
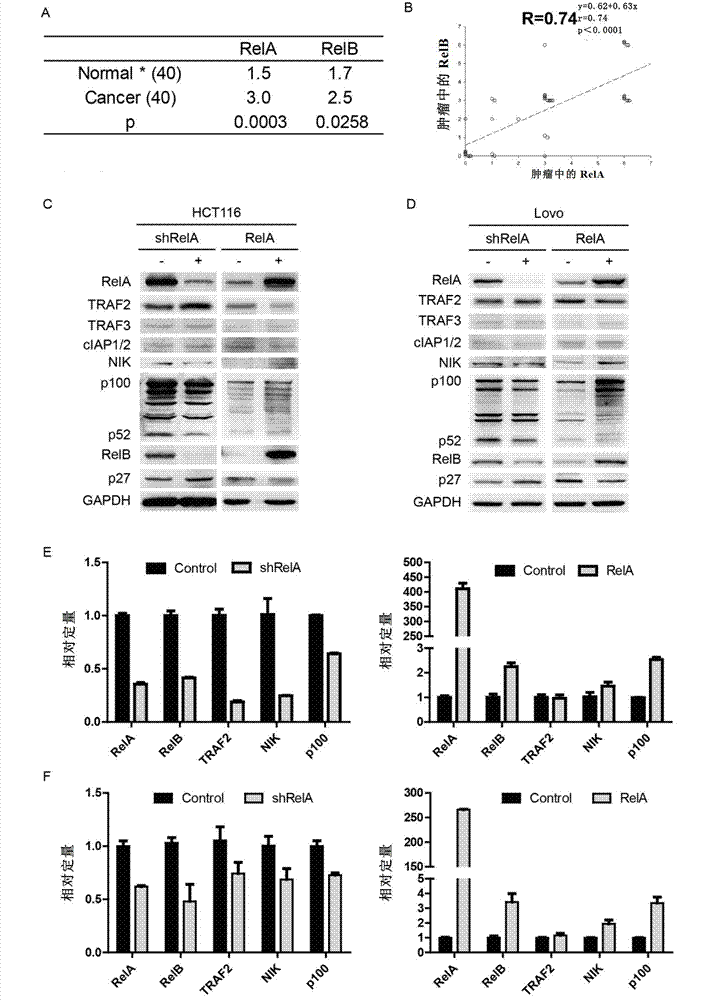
[0162] Canonical and non-canonical NF-κB pathways are constitutively activated in colorectal cancer
[0163] Test materials: Clinical tissue samples were obtained from Zhongshan Hospital Affiliated to Fudan University, and the shRelA vector was constructed with pLVX-shRNA1.
[0164] Test method: use Clontech's RNAi Designer tool to design the target shRNA. First use RNAi Target Sequence Selector to select the target sequence of shRNA, and then use shRNA Sequence Designer to design the corresponding shRNA sequence according to the target sequence. In order to facilitate the connection of shRNA to the vector, restriction enzyme sites were added at both ends of the shRNA sequence when designing the sequence. The commercially available plasmid vector pLVX-shRNA1 contains restriction sites for BamH1 and EcoR1, and restriction sites for these two enzymes were added to both ends of the designed shRNA sequence.
[0165] Targeting sequence:
[0166] 5'-GGTGCAGAAAGAGGACATT-3' (SEQ ID...
Embodiment 2
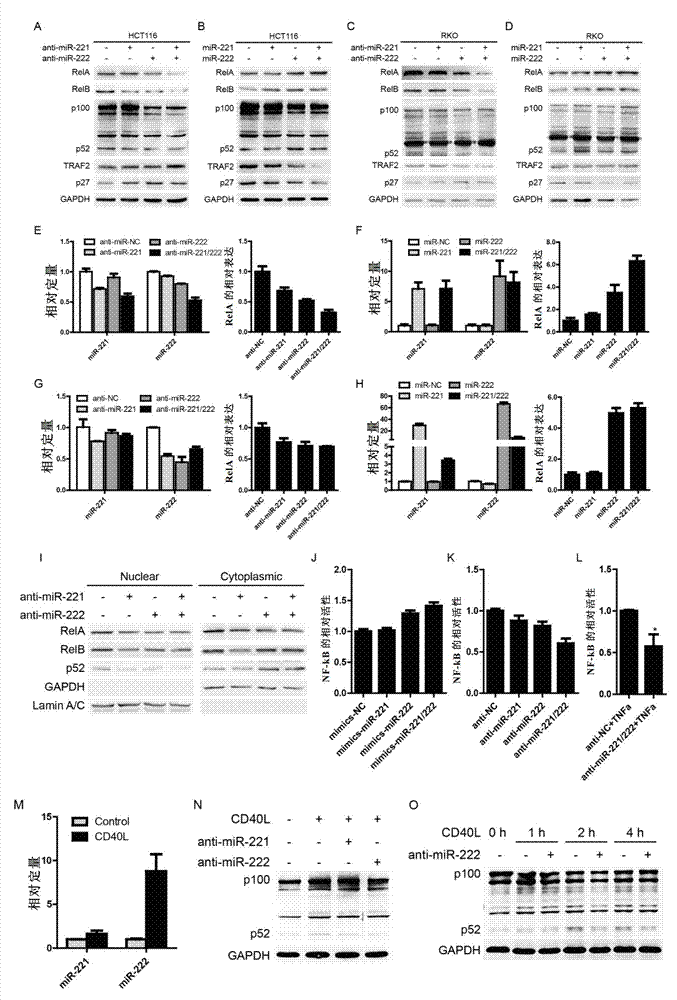
[0174] miR-221 / 222 affects the expression of RelA and affects the activation of canonical and non-canonical NF-κB pathways
[0175] Test materials: miR-221 / 222 mimics and antisense were purchased from Gemma. Real-time PCR kit was purchased from Takara Company. RNA inversion kit was purchased from Transgene. Dual fluorescent reporter gene detection kit was purchased from Promega.
[0176]Test method: 100nM miR-221 / 222 mimics or antisense were transfected into HCT116 and RKO cells, and 3 days after transfection, western blotting and real-time PCR were used to detect the expression of related genes. 100nM miR-221 / 222 mimics or antisense, 200ng NF-κB reporter gene vector and 50ngpRL-TK vector were co-transfected into HCT116 cell line, and 2 days after transfection, the activity of NF-κB reporter gene was detected. 1ng / ml CD40L stimulated HCT116 cells, collected samples after 12 hours, and detected the expression of miR-221 / 222. 100 nM of miR-221 / 222 was transfected into HCT116...
Embodiment 3
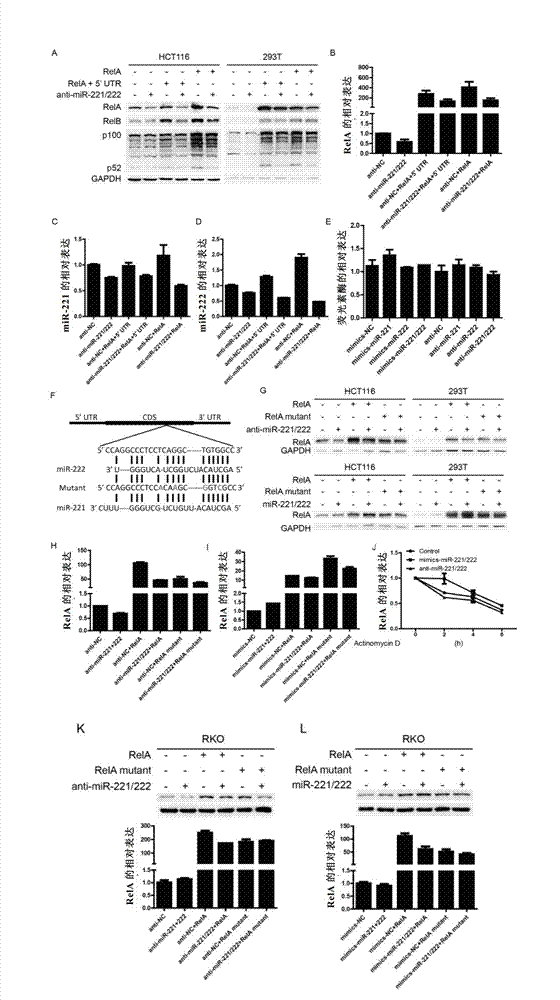
[0180] miR-221 / 222 affects the expression of RelA by regulating the coding region of RelA
[0181] Test materials: RelA expression vector was constructed with commercially available pcDNA3.1(+). The mutation kit was purchased from Takara Company. Actinomycin D was purchased from Shanghai Qianchen Biological Company.
[0182] Upper chain primer: 5'-ATA AAGCTT ATGGACGAACTGTTCCCCCTC-3' (SEQ ID NO.:12), lower strand primer: 5'-ATA GGATCC TTAGGAGCTGATCTGACTCAG-3' (SEQ ID NO.: 13), enzyme cutting sites: HindIII and BamHI.
[0183] Since miR-221 / 222 positively regulates the expression of RelA, the inventors tried to verify whether it promotes its expression by binding to the 5'UTR of RelA. Test method: 100nM miR-221 / 222 antisense and 2μg RelA expression vector were co-transfected into HCT116 or 293T cells, and 3 days after transfection, western blotting and real-time PCR were used to detect the expression of related genes. 100nM miR-221 / 222 mimics or antisense, and 200ng psiCH...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com