Method for rapidly detecting industrial saccharomyces pastorianus ester metabolism genes
A Pasteurella and gene technology, applied in the field of rapid detection of industrial Pasteurella ester metabolism genes, can solve the problem of low throughput, achieve strong specificity and sensitivity, improve typical flavor and consistency, and shorten experimental time Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
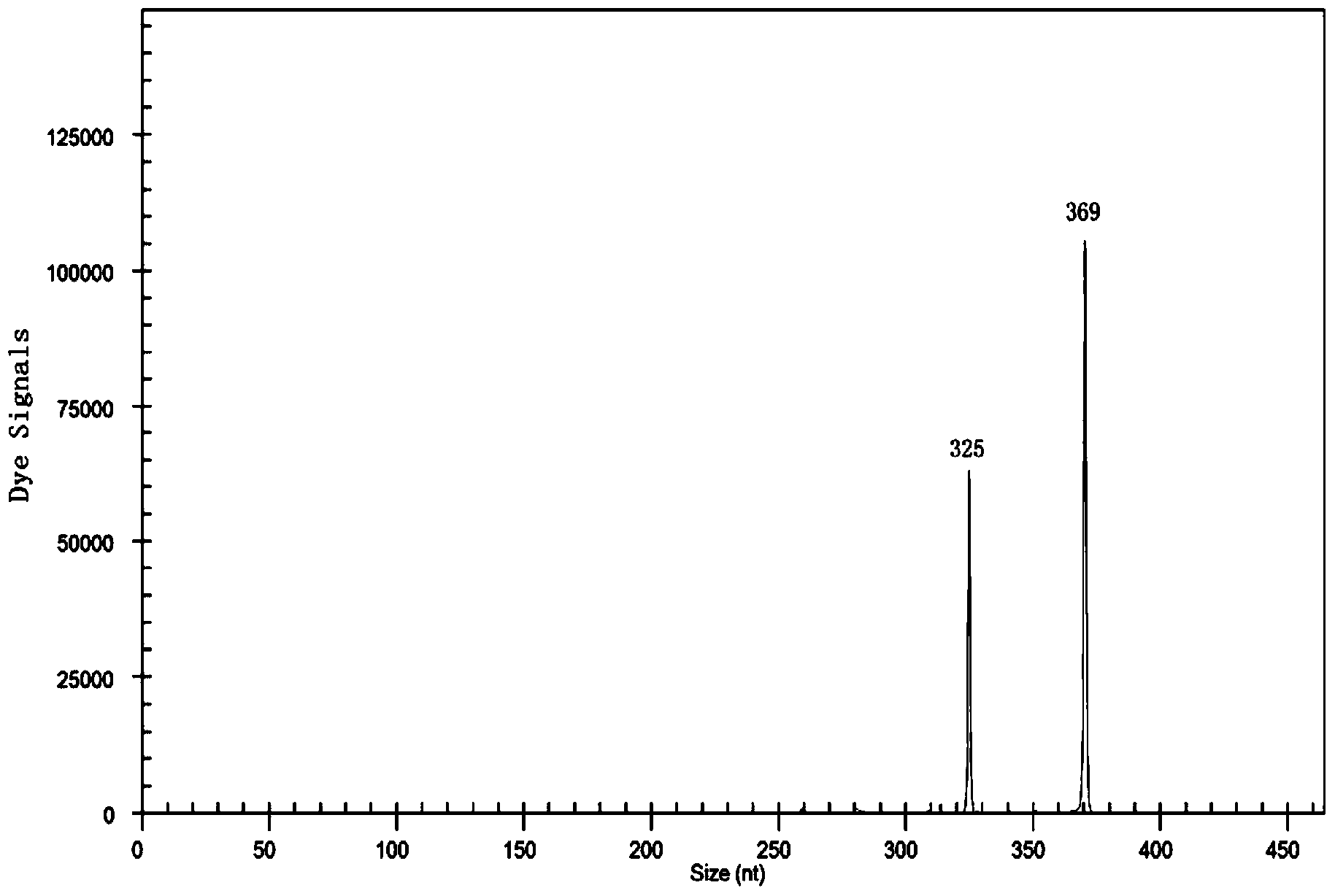
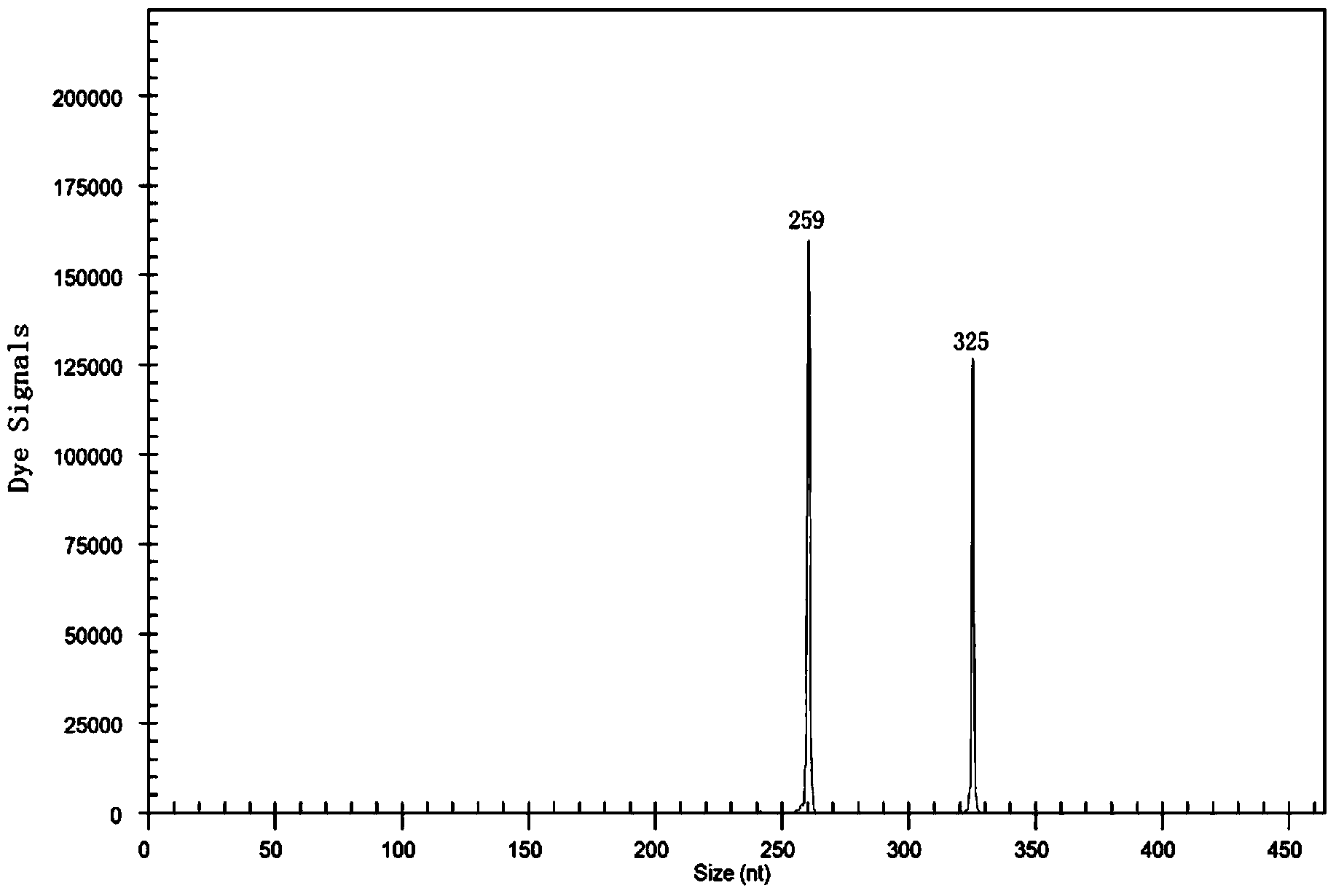
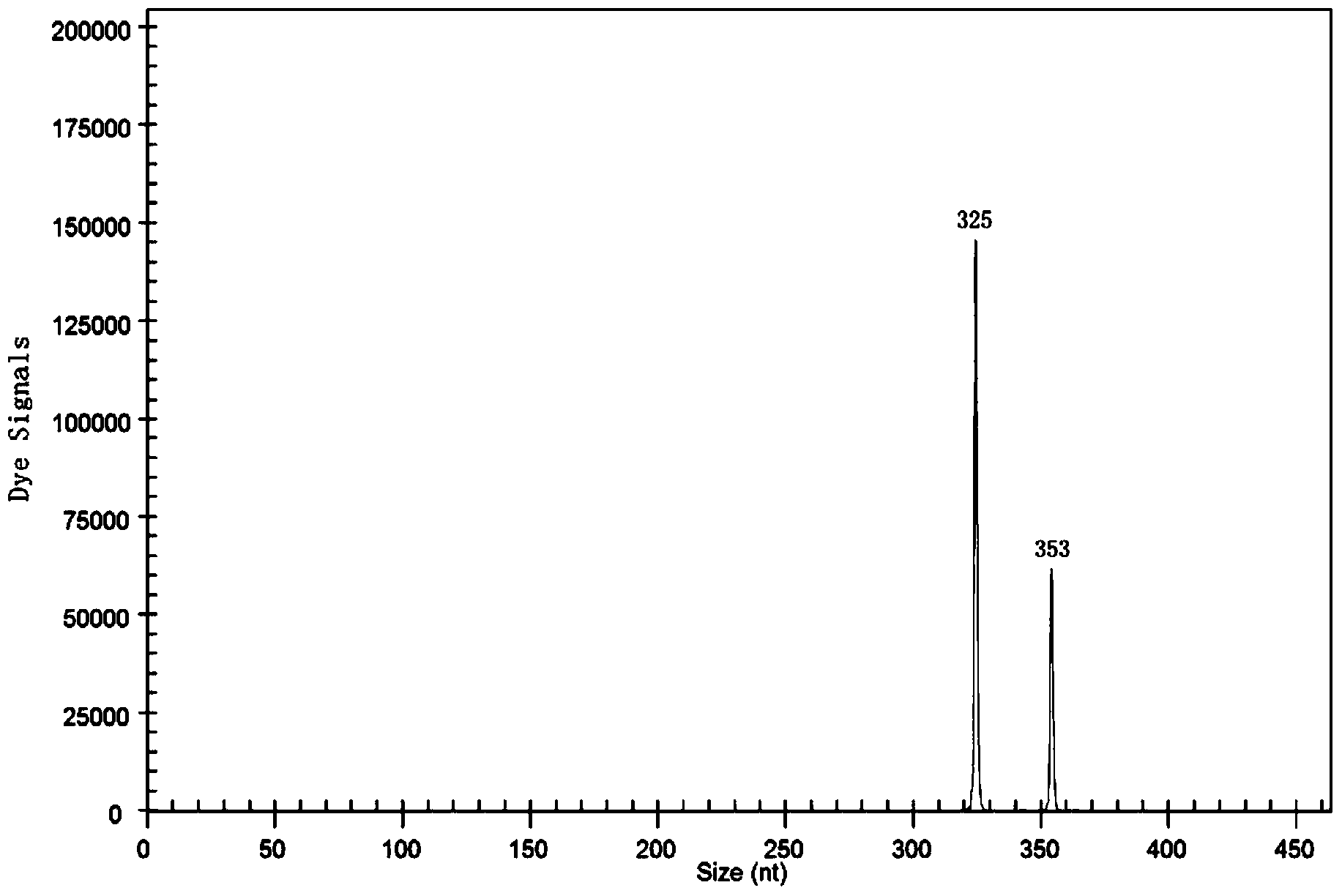
[0051] A method for rapidly detecting ester metabolism genes of Pasteurella industrialis, including extraction of total RNA, preparation of cDNA templates by reverse transcription, multiplex PCR reaction, capillary electrophoresis, and product fragment analysis, aimed at genes related to ester metabolism of Pasteurella industrialis ATF1-Sc, ATF1-Sb, ATF2-Sc, ATF2--Sb, EEB1-Sc, EEB1-Sb, EHT1-Sc, EHT1-Sb, IAH1-Sc, IAH1-Sb, reverse transcription reaction to prepare cDNA template application primers are SEQ ID NO.2, SEQ ID NO.4, SEQ ID NO.6, SEQ ID NO.8, SEQ ID NO.10, SEQ ID NO.12, SEQ ID NO.14, SEQ ID NO.16, SEQ ID NO.18, SEQ ID NO.20, primers SEQ ID NO.1, SEQ ID NO.3, SEQ ID NO.5, SEQ ID NO.7, SEQ ID NO.9, SEQ ID NO. 11. SEQ ID NO.13, SEQ ID NO.15, SEQ ID NO.17, SEQ ID NO.19. It also includes industrial Pasteurella β-actin genes ACT1-Sc and ACT1-Sb as internal reference genes. The primers used to prepare cDNA templates in reverse transcription reactions are SEQ ID NO.22 and SEQ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com