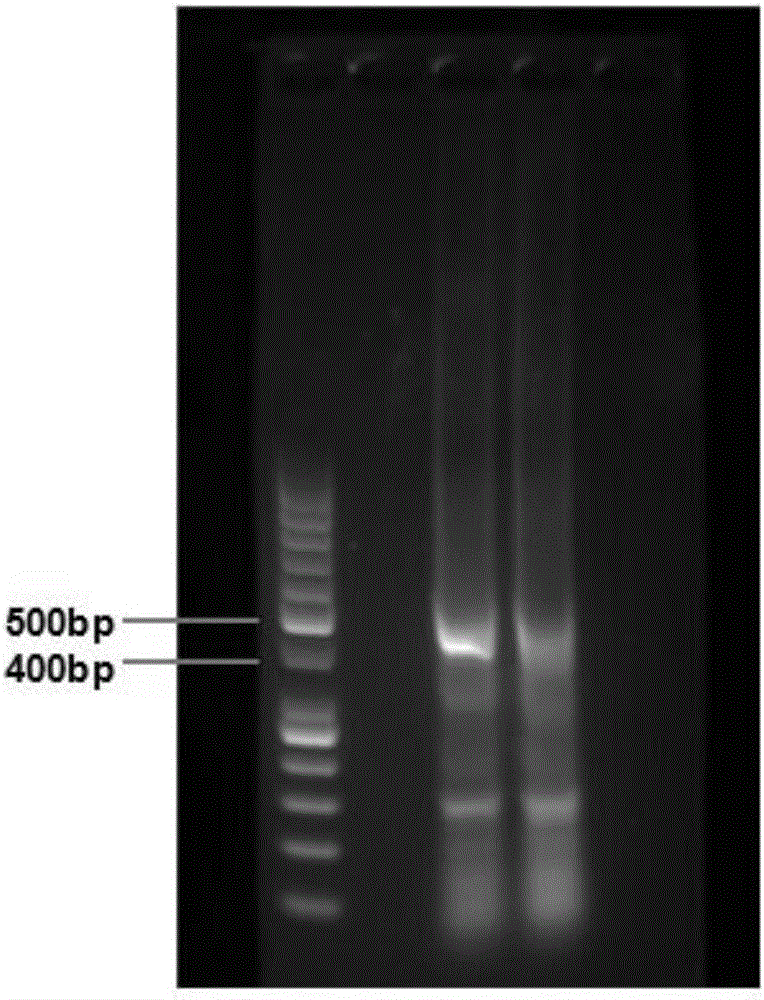
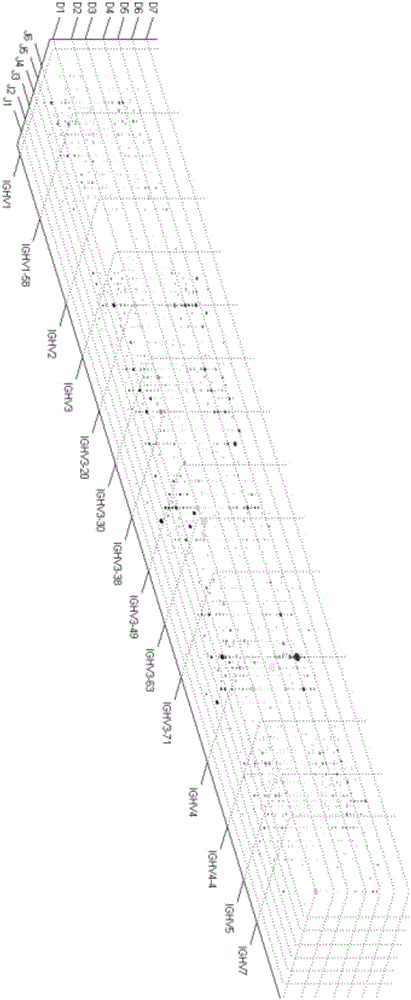
Method for high-throughput sequencing of tcr or bcr and method for correcting multiple pcr primer bias by using tag sequence
A tag sequence, high-throughput technology, applied in the field of molecular biology, can solve the problems of PCR primer amplification error, increase the complexity of primers, etc., and achieve the effect of reducing sequencing error, length and quantity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0075] Embodiment 1 of the present invention provides a method for preparing a B lymphocyte receptor (BCR) RNA sample, comprising the following steps:
[0076] (1) Collect 10 milliliters (ml) of fresh peripheral blood samples from healthy people;
[0077] (2) Using the principle of density gradient centrifugation (the density of Ficoll is 1.077, and the average density of PBMC is 1.072), and through repeated centrifugation and washing, relatively pure PBMCs are obtained. The steps are as follows:
[0078] a. Dilute the blood obtained in step (1) 1:1 with PBS or 0.09% NaCl;
[0079] b. Add 0.5 times the volume of the resulting diluted blood lymphocyte separation solution to the 15ml centrifuge tube;
[0080] c. Slowly add the diluted blood obtained in step a to the surface of the lymphocyte separation liquid obtained in step b to keep the separation liquid and blood layered, and then centrifuge with a swingangle centrifuge at a speed of 600g for 20min-25min at a lower speed, a...
Embodiment 2
[0085] Embodiment 2 of the present invention provides a method for using BCR full-length primers to perform two cycles of multiplex PCR to construct a BCR full-length high-throughput sequencing library for high-throughput sequencing, including the following steps:
[0086] (1) Reverse transcribe the total RNA obtained in Example 1 into cDNA, the method is as follows:
[0087] (1.1) Configure the following system on ice:
[0088]
[0089]
[0090]The BCR full-length downstream primers in this system are mIgG, mIgA, mIgM, mIgE or mIgD downstream primers (respectively shown in SEQ ID NO: 23 ~ SEQ ID NO: 27) and mixed with sequencing adapters, specifically as SEQ ID NO: 23 The 5' ends of the primers shown in ~SEQ ID NO:27 were respectively connected with the downstream primer adapter sequence of Illumina Sequencing Company and then mixed equimolarly. Throughput sequencing library construction instruction), the total RNA is the total RNA prepared in Example 1, and the water ...
Embodiment 3
[0114] Example 3 of the present invention provides a method of using CDR3 primers of BCR to perform two cycles of multiplex PCR to construct a high-throughput sequencing library in the CDR3 region of BCR for high-throughput sequencing. The full length of -V H 1. Full length-V H 2. Full length-V H 3. Full length-V H 4. Full length-V H 5. Full length-V H 6 and full length-V H The upstream primer of 7 was replaced by CDR3-V H 1. CDR3-V H 2. CDR3-V H 3. CDR3-V H 4. CDR3-V H 5. CDR3-V H 6 and CDR3-V H 7 upstream primers (as shown in SEQ ID NO: 28 to SEQ ID NO: 39), other operations and experimental conditions remained unchanged.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com