Library versus library yeast two-hybrid massive interaction protein screening method
A yeast two-hybrid, protein technology, applied in the field of molecular biology, can solve problems such as incompatibility, and achieve the effects of improving efficiency, low hardware requirements, and simple operation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
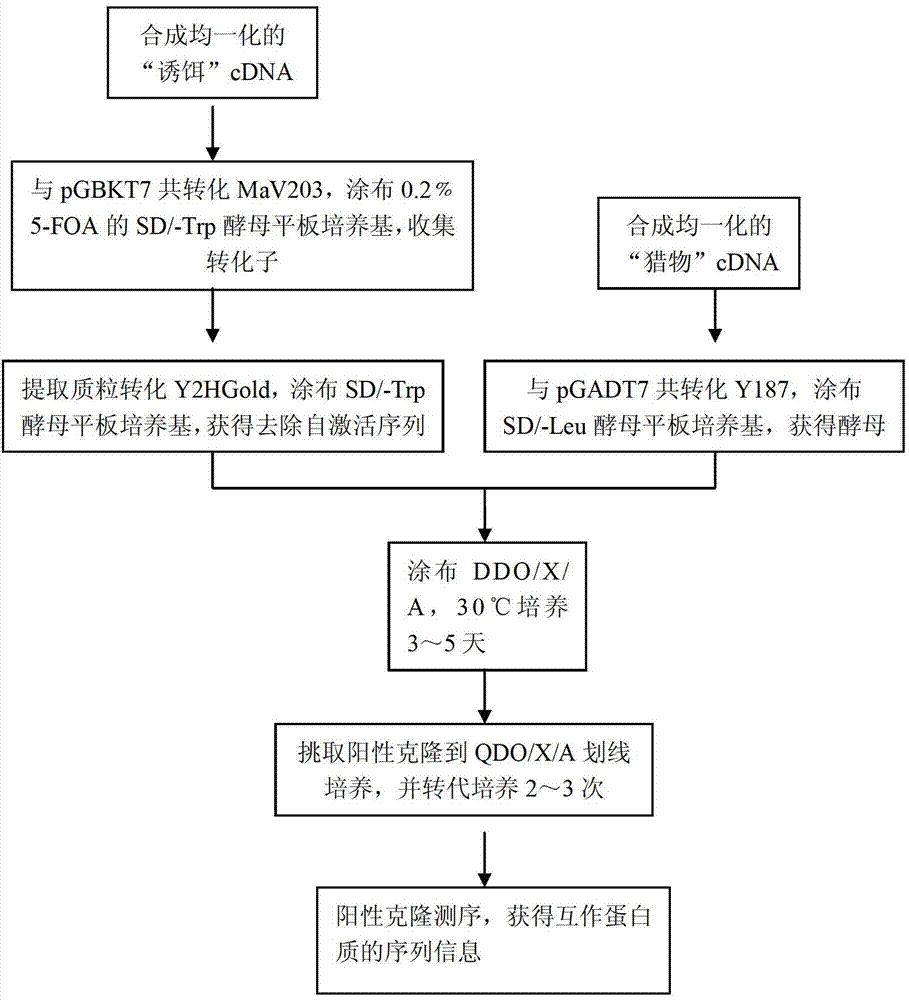
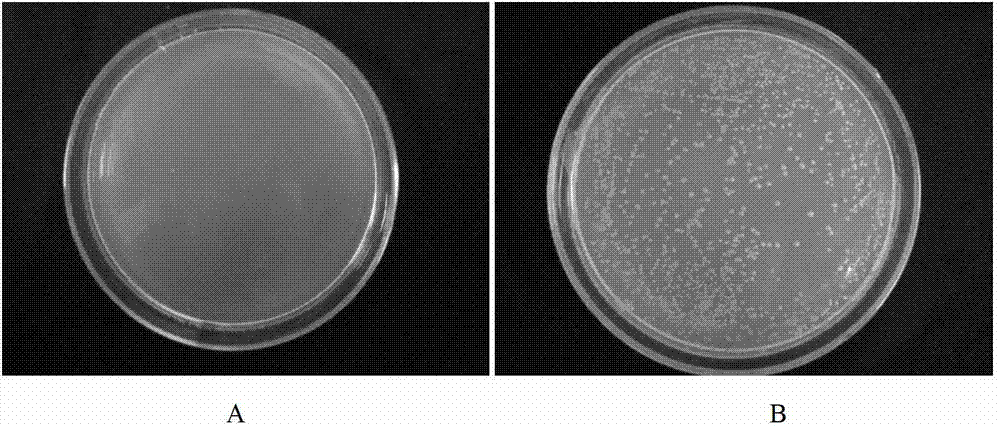
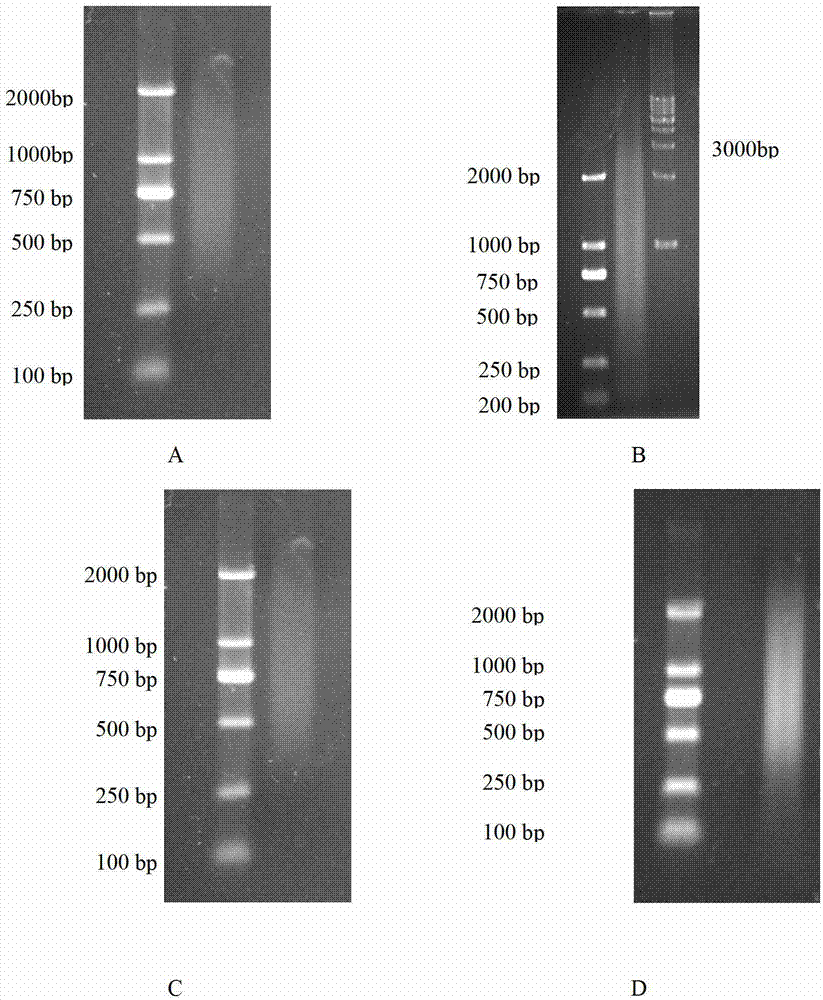
[0045] Example 1: A library-to-library yeast two-hybrid method for large-scale screening of interacting proteins: Screening of proteins interacting between the plant pathogen Rhizoctonia solani AG Ⅰ-IA and the host corn, the specific steps are as follows:
[0046] (1) Synthesize the "bait" cDNA-Rhizoctonia solani AG Ⅰ-IA cDNA and the "prey" cDNA-corn cDNA respectively:
[0047] a. Trizol reagent from Invitrogen Company extracted the total RNA of Rhizoctonia solani AG Ⅰ-IA and corn leaves inoculated and infected with Rhizoctonia solani AG Ⅰ-IA, and purified the mRNA. -IA mRNA 323.2 ng / μl, maize mRNA 309.6 ng / μl.
[0048] b. Add the following reagents to the RNAase-free microcentrifuge tube in order
[0049] mRNA
1.5μl
GBK-CDS Ⅲ (Rhizoctonia solani AG Ⅰ-IA) / CDS Ⅲ (corn)
1μl
RNAase-free sterile water
1.5μl
total capacity
4μl
[0050] Mix well and centrifuge to the bottom of the tube.
[0051] Primer GBK-CDS Ⅲ was synthesize...
Embodiment 2
[0103] Example 2 A library-to-library yeast two-hybrid method for large-scale screening of interacting proteins: screening of interacting proteins between plant pathogenic bacteria powdery mildew of wheat and host wheat, the specific steps are as follows:
[0104] (1) Separately synthesize "bait" cDNA-wheat powdery mildew cDNA and "prey" cDNA-wheat cDNA:
[0105] a. Trizol reagent from Invitrogen Company extracted the total RNA of wheat powdery mildew bacteria, wheat leaves inoculated and infected with wheat powdery mildew bacteria, and purified to obtain mRNA.
[0106] b. Add the following reagents to the RNAase-free microcentrifuge tube in order
[0107] mRNA
1.0μl
GBK-CDS Ⅲ (wheat powdery mildew) / CDS Ⅲ (wheat)
1.0μl
RNAase-free sterile water
2.0μl
total capacity
4.0μl
[0108] Mix well and centrifuge to the bottom of the tube.
[0109] c. Incubate at 72°C for 2 minutes, cool on ice for 2 minutes, centrifuge to the bott...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com