Method for rapid identification of microalgae on single cell level
A single-cell, microalgae technology, used in material excitation analysis, Raman scattering, etc., to avoid errors and reduce sample requirements
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
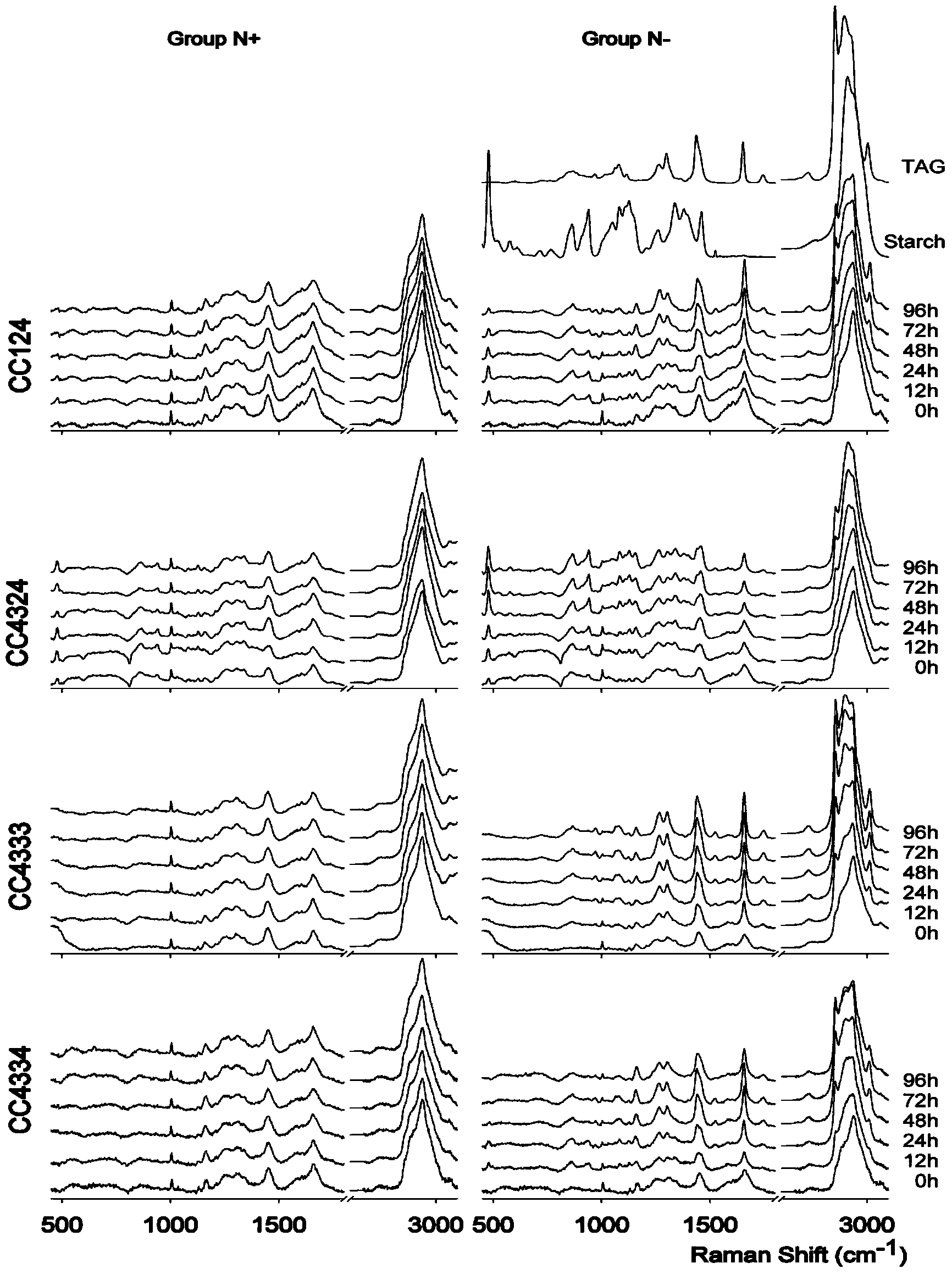
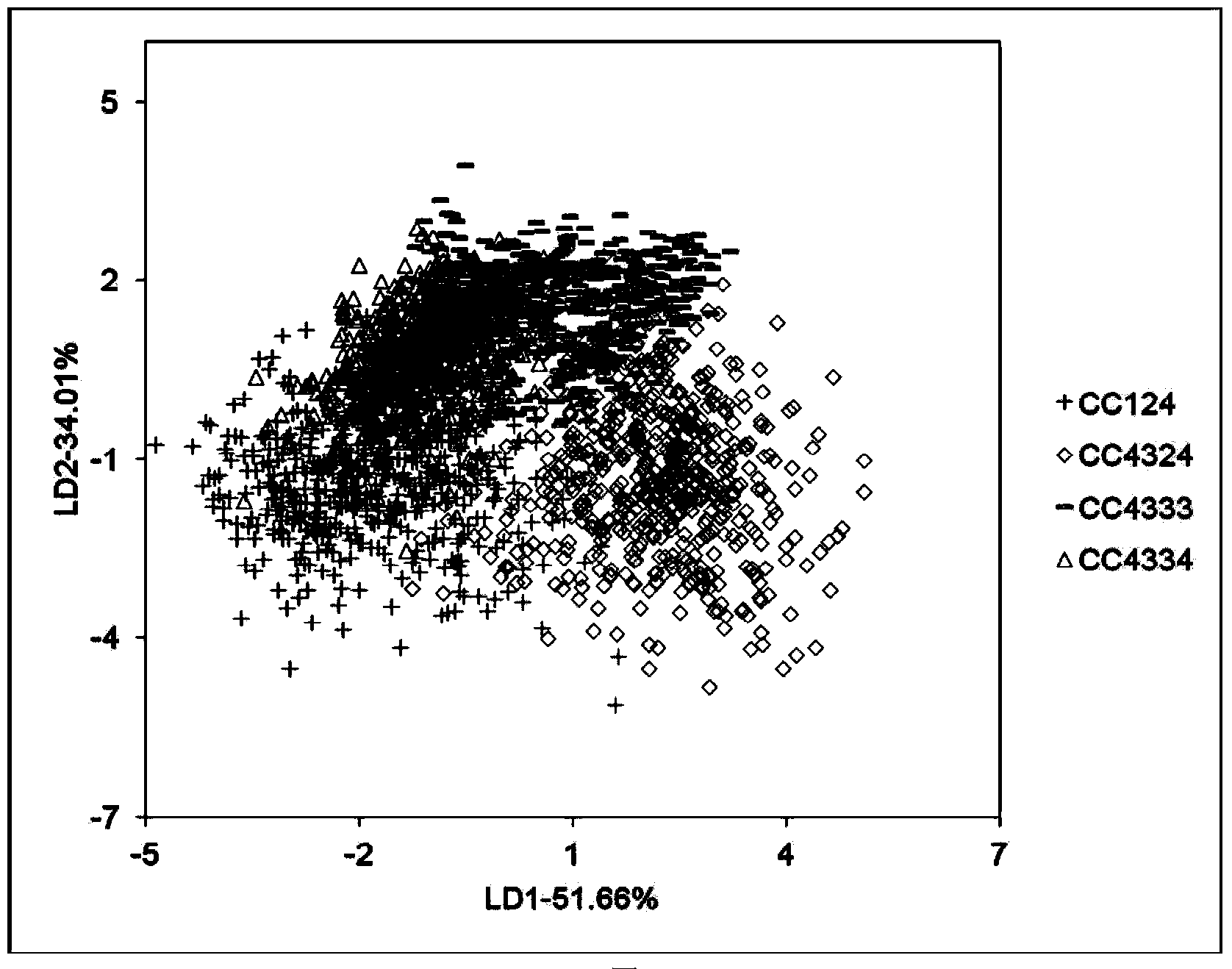
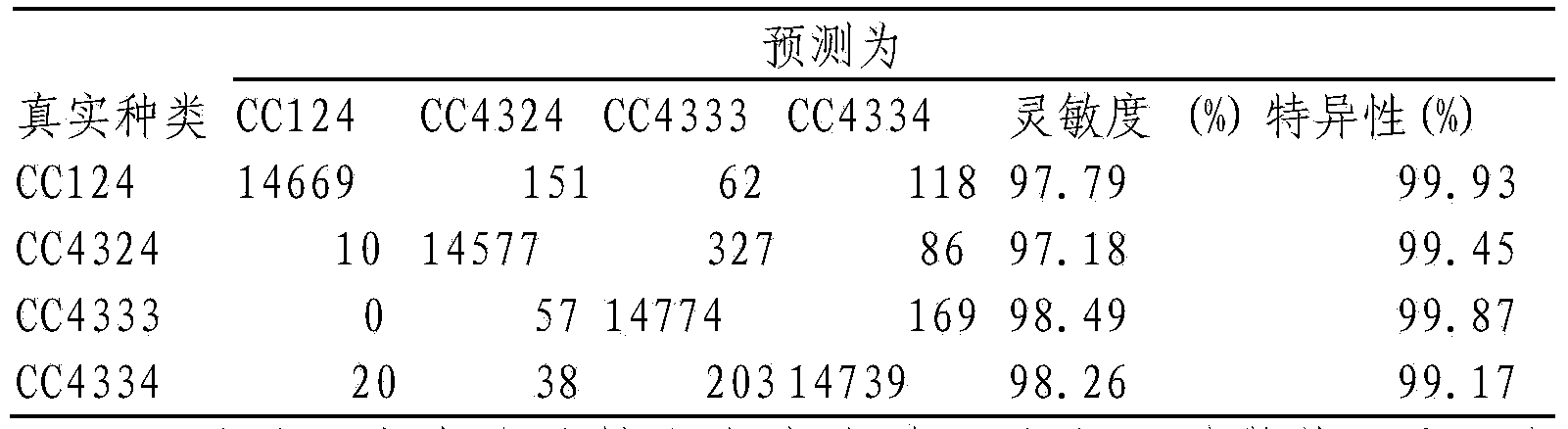
[0024] 1) Four strains of Chlamydonomas reinhardtii mutants CC124, CC4324, CC4333 and CC4334 were purchased from Chlamydomonas Resource Center: http: / / chlamycollection.org / strains / .
[0025] 2) Chlamydomonas reinhardtii was inoculated into TAP (Tris acetate phosphate) liquid medium with or without nitrogen (3 biological replicates), 150 μmol photons m -2 the s -1 Light, 25 ° C, aeration and stirring culture. Samples were taken when culturing to 0h, 12h, 24h, 48h, 72h and 96h. Among them, TAP liquid medium with nitrogen or nitrogen deficiency, TAP (Tris acetate phosphate) medium:
[0026] NH 4 Cl (7.48mM), MgSO 4 (406 μM), CaCl 2 (340μM), K 2 HPO 4 (540μM), KH 2 PO 4 (463μM), 20mM Tris, 17.4mM acetate, H 3 BO 3 (184μM), ZnSO 4 (76.5μM),MnCl 2 (25.5μM), FeSO 4 (17.9 μM), CoCl 2 (6.77μM),(NH4) 6 Mo 7 o 24 (0.88μM), CuSO 4 (6.29μM), and Na 2 EDTA (148μM)
[0027] 3) Take the algae liquid of different culture time in step 2), take 1mL of the algae liquid for ea...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com