Haploid cells
A haploid cell, haploid technique, applied in the field of newly identified genes
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0213] [Example 1: Method]
Embodiment 11
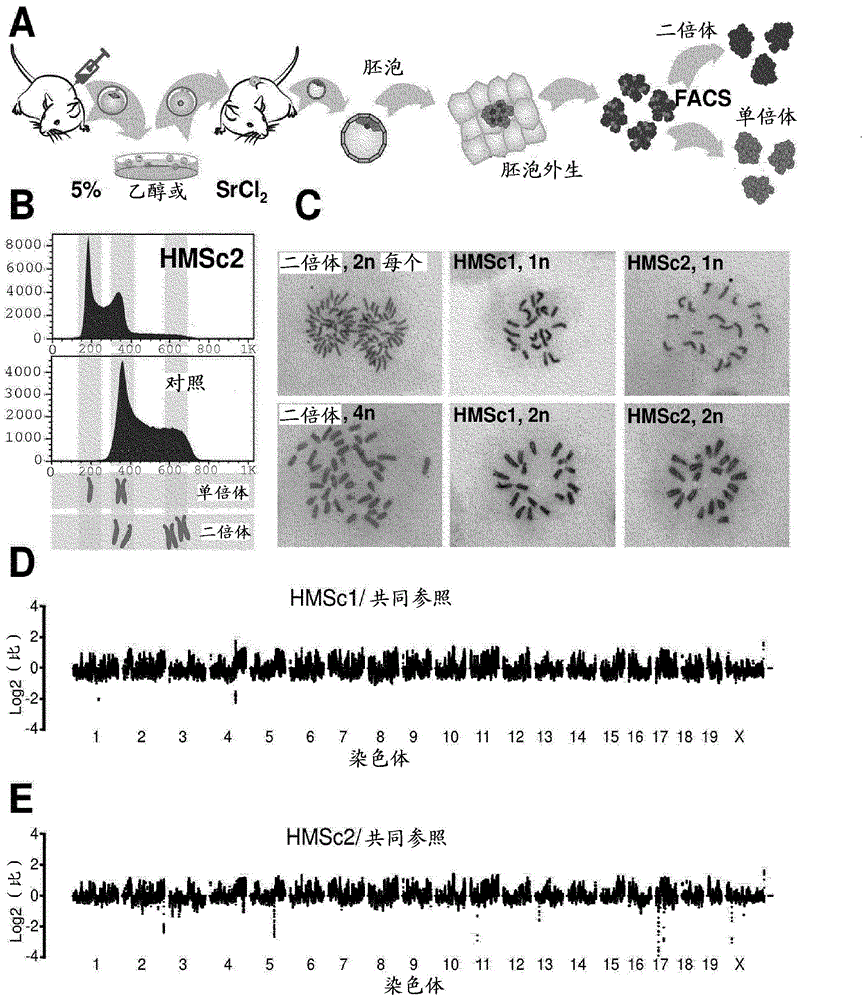
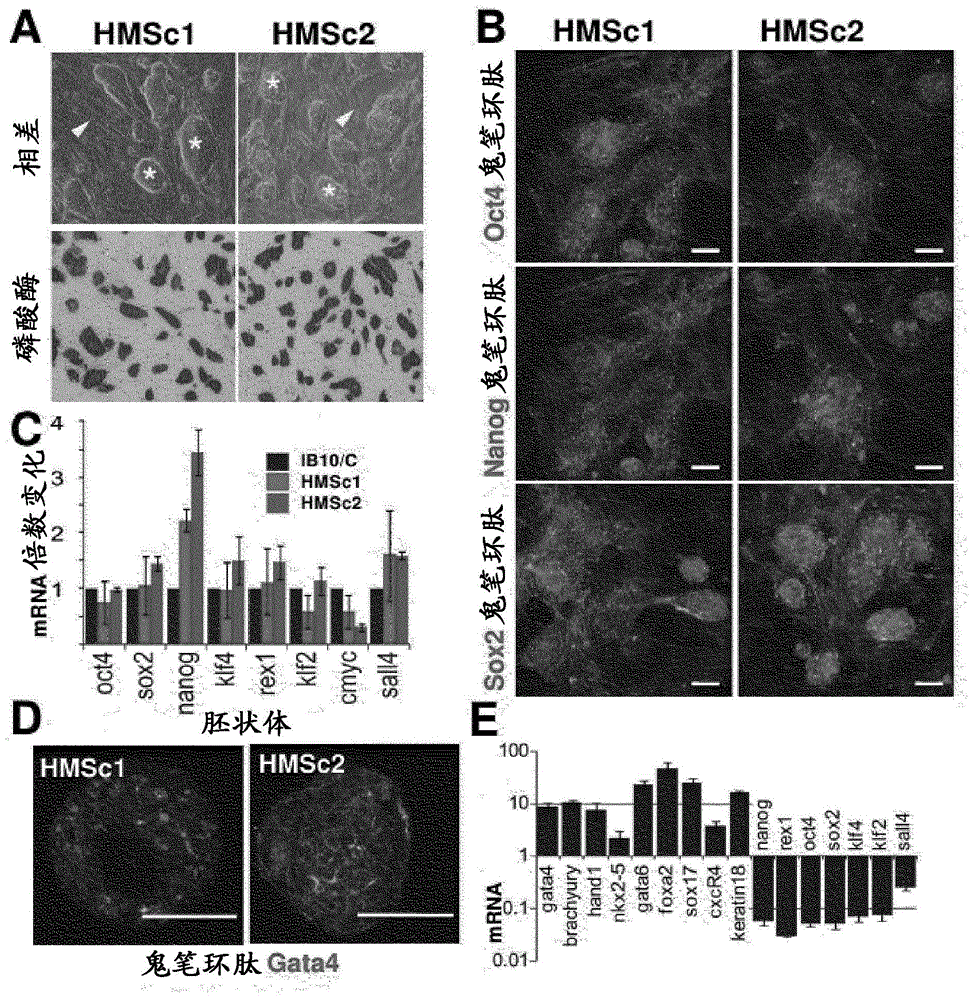
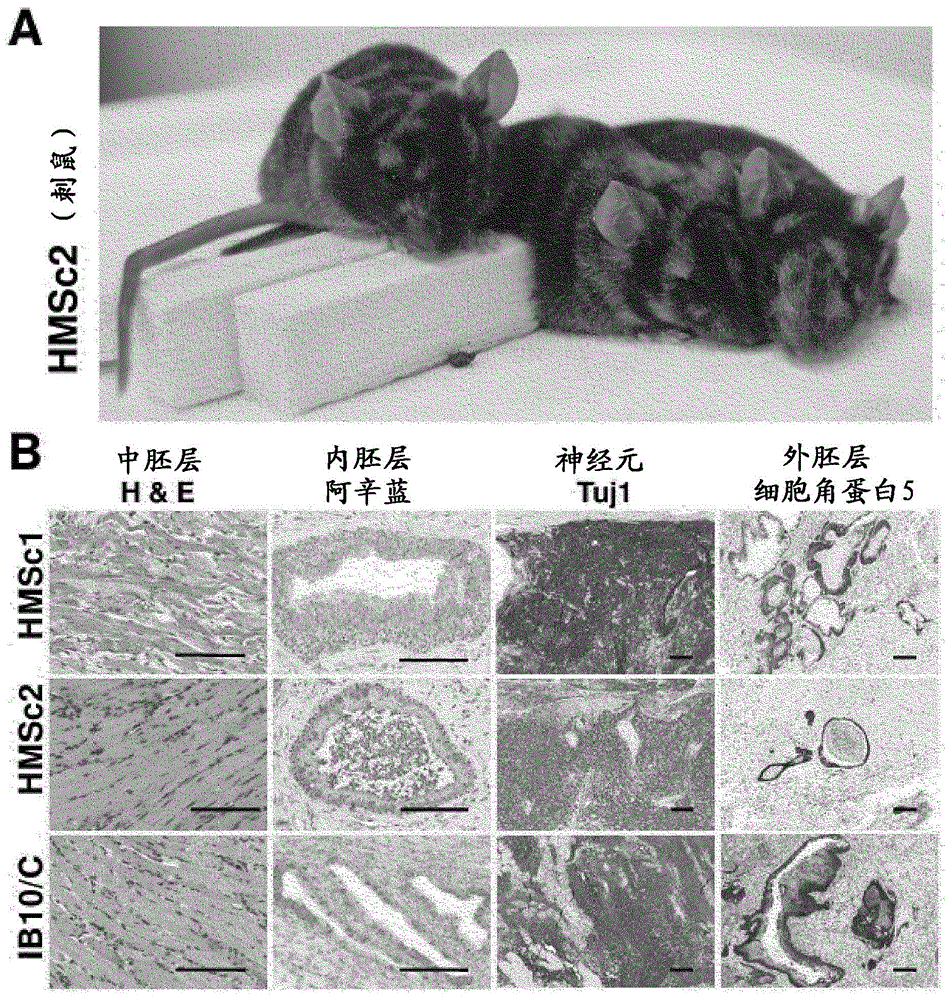
[0214] [Example 1.1: Parthenogenetic Derivation of Haploid Stem Cells]
[0215] C57BL / 6 x 129F1 female mice were superovulated using standard procedures, and unfertilized oocytes were washed and collected. For activation, oocytes were exposed to 5% ethanol or 25 mM SrCl as described 2 (Kaufman et al., 1983; Otaegui et al., 1999). After 4 hours of activation, viable oocytes were transferred into pseudopregnant 129 females, recollected at embryonic day 3.5 (ED), and cells were derived according to established embryonic stem cell derivation protocols (Bryja et al., 2006). Parthenogenetically derived stem cells are initially maintained on feeder layers and subsequently adapted to feeder-free culture conditions. Haploid cells are trained to grow under feeder-free conditions by gradually reducing the feeder cell density and eventually removing the feeder cells from the culture. Stem cell medium consisted of DMEM with 15% FCS (Gibco) supplemented with 2 mM L-glutamate, 1 mM sodium...
Embodiment 12
[0216] [Example 1.2: Genome coverage analysis and SNP mapping]
[0217] Genomic DNA preparations were sheared, adapters ligated and subjected to Illumina sequencing (HiSeq) by using a Covaris DNA sonicator according to the manufacturer's protocol. Only the Unique genome coordinates. SNPs, which differ between C57BL / 6 and 129, were retrieved from Sanger (www.sanger.ac.uk / resources / mouse / genomes / ) and mismatches observed during genomic mapping of deep sequencing reads against them were evaluated. Each SNP covered by Solexa reads was assigned to C57BL / 6 and 129 based on the majority of reads.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com