Method for improving expression amount of secretory foreign protein in pichia pastoris
A Pichia pastoris and exogenous protein technology, applied in the field of genetic engineering, can solve the problems of low secretion and expression of exogenous proteins and large differences in expression, and achieve the effect of increasing expression, improving correct folding, and promoting wide application
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1p
[0028] Example 1 Construction of pGAPZA and pGAPZB plasmids
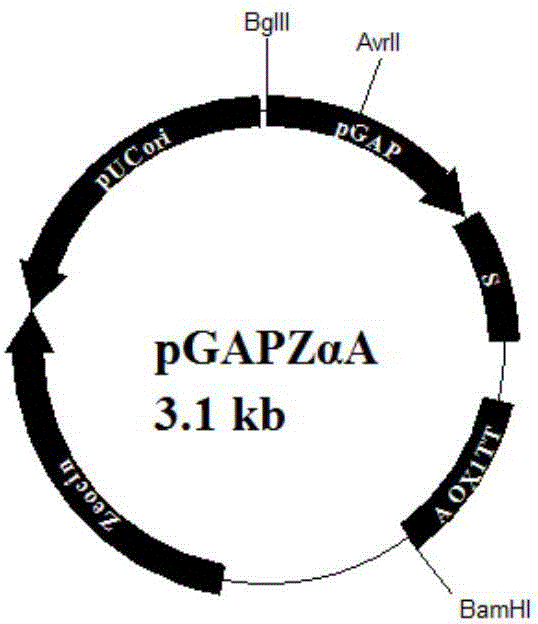
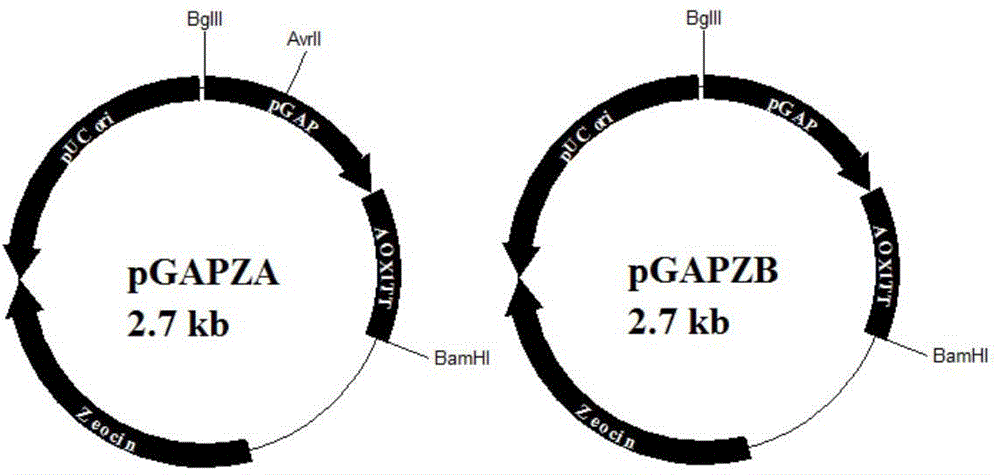
[0029] Plasmid pGAPZαA (purchased from Invitrogen) ( figure 1 ) with α-factor signal peptide, suitable for secreting and expressing exogenous protein, when transforming Pichia pastoris host, it is firstly digested with AvrII enzyme through the AvrII restriction site and then linearized, and the linearized fragment can be better integrated into the host genome superior. The regulatory factors HAC1, ERO1, and BIP need to be expressed intracellularly, so it is necessary to transform the plasmid pGAPZαA, remove the signal peptide, and construct a new plasmid pGAPZA ( figure 2 ), and on the basis of plasmid pGAPZA, the restriction enzyme site AvrII was mutated to construct a new plasmid pGAPZB.
[0030] The specific construction process is as follows:
[0031] The gene fragments of promoter pGAP and terminator AOX1TT were cloned by PCR reaction, and then the two fragments were fused together by overlapping PCR reacti...
Embodiment 2
[0041] Example 2 HAC1, ERO1, BIP gene amplification
[0042] The nucleotide sequences of HAC1, ERO1, and BIP genes were obtained from the KEGG database, which are SEQ ID NO: 1, SEQ ID NO: 2, and SEQ ID NO: 3, respectively. These three genes were amplified using the primers described below.
[0043] Primer 7 (HAC1-F): ATGCCCGTAGATTCTTCTC
[0044]Primer 8 (HAC1-R): TCACCTGATCGCTATGCAT
[0045] Primer 9 (ERO1-F): ATGAGGATAGTAAGGAGCG
[0046] Primer 10 (ERO1-R): TTACAAGTCTACTCTATAT
[0047] Primer 11 (BIP-F): ATGCTGTCGTTAAAACCAT
[0048] Primer 12 (BIP-R): CTACAACTCATCATGATCA
[0049] Using the genome of Pichia pastoris GS115 as the template, primers 7 and 8 were amplified to obtain the HAC1 gene with a size of 996bp; primers 9 and 10 were amplified to obtain the ERO1 gene with a size of 1584bp; primers 11 and 12 were amplified to obtain the BIP gene with a size of 2037bp ; The 3 fragments were connected to T vector and sequenced, and the sequence was consistent with the seq...
Embodiment 3
[0050] Example 3 Construction of pGAPHE and pGAPHEB plasmids
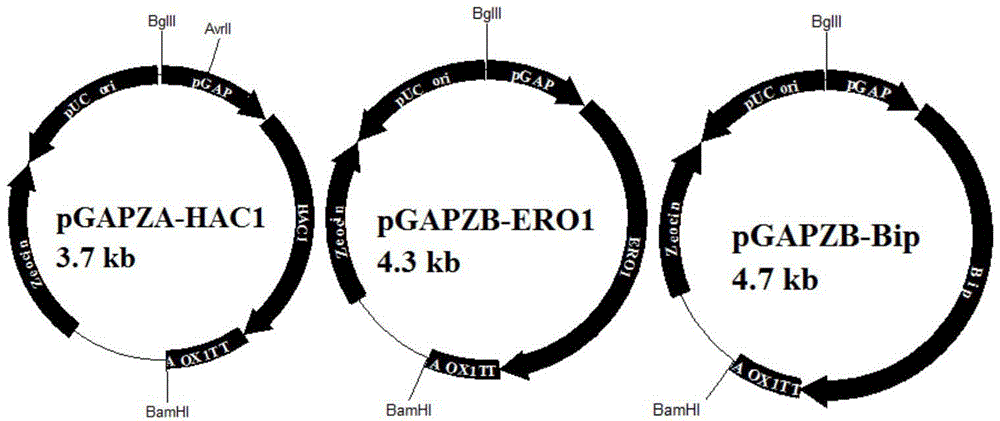
[0051] EcoR I and Not I restriction sites were introduced at both ends of the HAC1, ERO1 and BIP gene fragments by PCR. After double restriction digestion, HAC1 was connected to the pGAPZA plasmid, and ERO1 and BIP were connected to the pGAPZB plasmid to construct pGAPZA-HAC1 and pGAPZB-ERO1 respectively. , pGAPZB-BIP plasmid ( image 3 ).
[0052] After the pGAPZB-ERO1 plasmid was double digested with Bgl II and BamH I, the expression cassette containing the promoter, the target gene and the terminator was recovered by gel, and the size was 2486bp. After the pGAPZA-HAC1 plasmid was digested with BamH I, it was ligated with the expression cassette, transformed into the DH5α strain, and the positive transformants were picked. Since the digestion sites of Bgl II and BamHI are homo-caudal enzymes and have the same sticky ends, two plasmids, forward and reverse, will be formed when ligating, which can be verified by ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com