Method for detecting trinucleotide repeated sequence of mammal genome and application thereof
A technology of trinucleotides and repeating sequences, applied in the field of nucleotide detection, can solve problems such as inability to carry out specific analysis
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
specific example
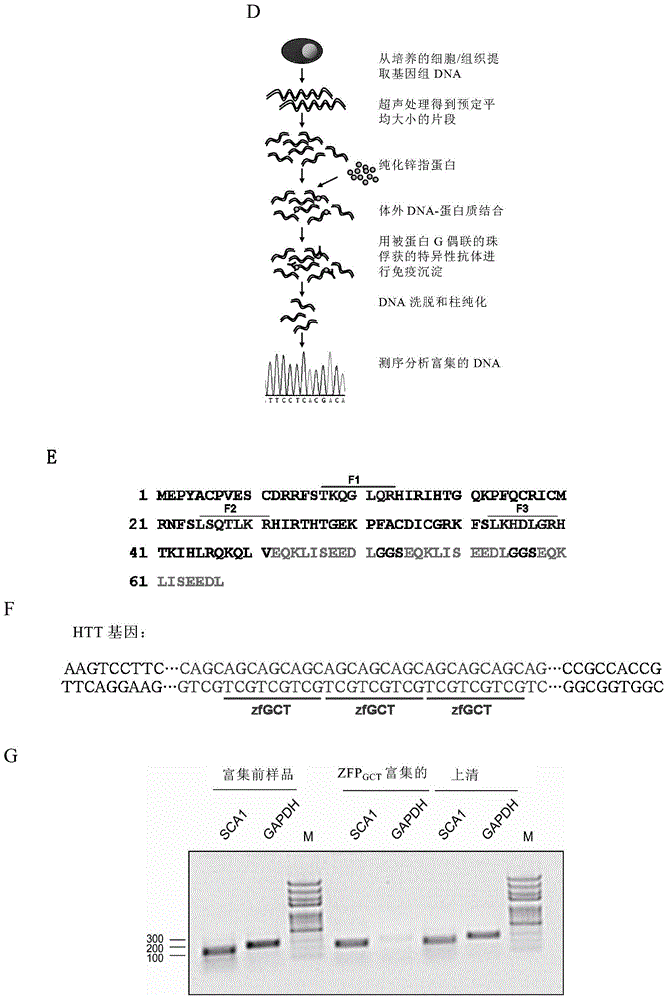
[0064] As a specific example, the method includes:
[0065] (1) incubating the special sequence binder and the DNA under conditions suitable for the binding of the special sequence binder to the DNA;
[0066] (2) Isolate the bound specific sequence binder-DNA complex;
[0067] (3) DNA obtained by separating and purifying the special sequence binder-DNA complex obtained in step (2); and
[0068] (4) Sequencing the DNA separated and purified in step (3).
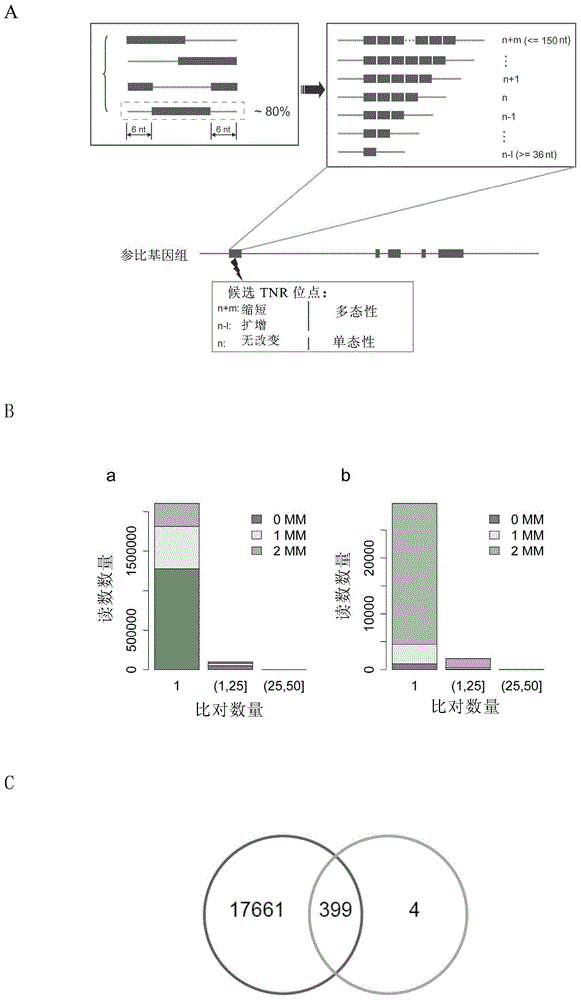
[0069] Herein, special sequences include, but are not limited to, trinucleotide repeat sequences, including (GCT)n, (CTG)n, (TGC)n, (AGC)n, (GCA)n, (CAG)n, (GGC) n, (GCG)n, (CGG)n, (GCC)n, (CCG)n, (CGC)n, (GAA)n, (AAG)n, (AGA)n, (TTC)n, (TCT)n n and (CTT)n, where n is an integer greater than or equal to 3. In a specific embodiment, n≤100, for example, n is an integer between 3-50, 3-30, and 3-20.
[0070] Herein, the specific sequence combination includes zinc finger protein or transcriptional activator-like effector, and...
Embodiment 1
[0155] Example 1: Specific binding of recombinant zinc finger protein to target DNA
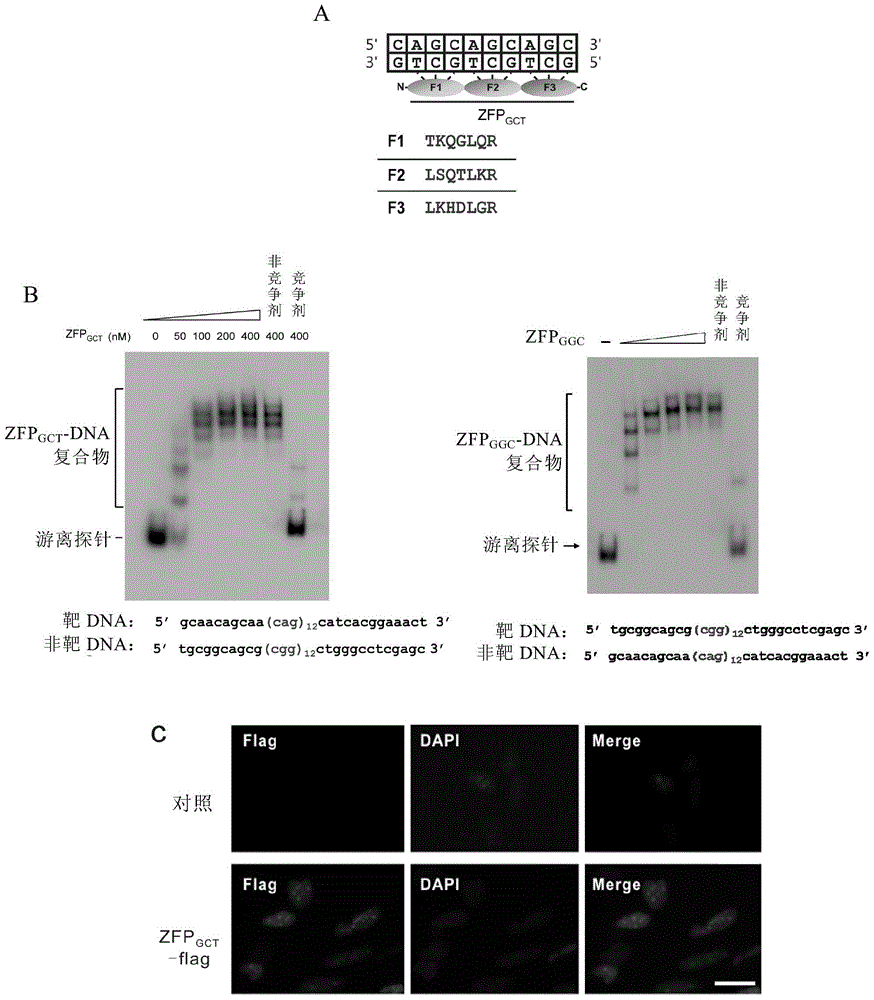
[0156] We designed a specific recognition GCT trinucleotide repeat sequence (GCT) based on the zif268 zinc finger protein 3 zinc finger protein ZFP GCT (SEQ ID NO:1), which is connected by three tandem zinc finger domains, in which the specific amino acid residues that recognize the helix are in figure 1 listed in A. We constructed the artificially designed recombinant gene on the E.coli expression vector, and purified it with NTA-Ni column after inducing expression.
[0157] We used the same method to prepare the zinc finger protein ZFP GGC (SEQ ID NO:2) and ZFP GAA (SEQ ID NO: 3).
[0158] At the same time, we constructed the recombinant gene into a eukaryotic cell expression vector, transfected it into HeLa cells through liposomes, and verified the localization of the protein in eukaryotic cells by immunofluorescence. By co-staining with DAPI dye, we can see the ZFP GCT The -flag fu...
Embodiment 2
[0162] Example 2: ZFPs GCT Binds specifically to CTG, TGC, GCT, AGC, GCA and CAG repeats
[0163] We used EMSA experiments and in vitro ZIP experiments (ie, the aforementioned in vitro DNA immunoprecipitation) to prove that ZFP GCT Protein specific binding to GCT repeats. Such as figure 1 As shown in F, there are multiple CAG repeats in the HTT gene, each of which has a ZFP GCT The protein can bind three GCT repeats, so HTT genomic DNA can be read by multiple ZFPs GCT protein binding. When ZFP GCT When binding to a sequence containing ≥ 3 repeated GCTs, the reading frame is shifted, and it can also specifically bind to CTG, TGC and AGC, GCA, CAG repeat sequences of the complementary chain. At the same time, the method of semi-quantitative PCR was used to detect the DNA enriched by ZIP in vitro, such as figure 1 As shown in G, compared with the non-specifically enriched GAPDH gene fragment, the specifically enriched SCA1 gene fragment can be significantly amplified.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com