Sanger sequencing reaction optimization agent, sequencing reaction system and sequencing method employing optimization agent
A sequencing reaction system and sequencing reaction technology, applied in the field of molecular biology, can solve the problems of short effective sequencing sequences, interrupted sequencing signals, and inaccurate sequences.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
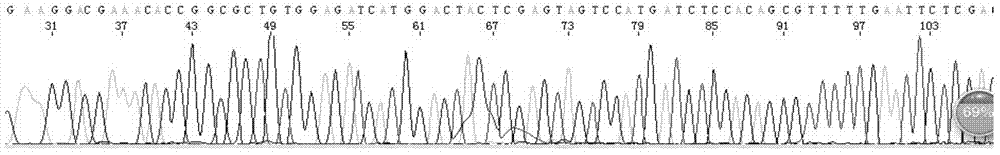
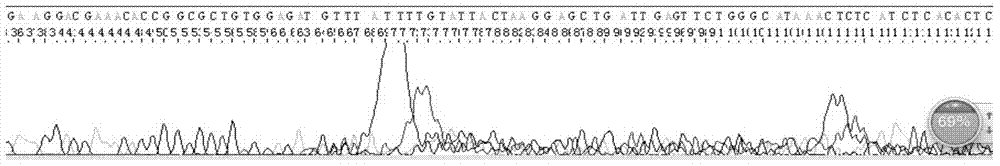
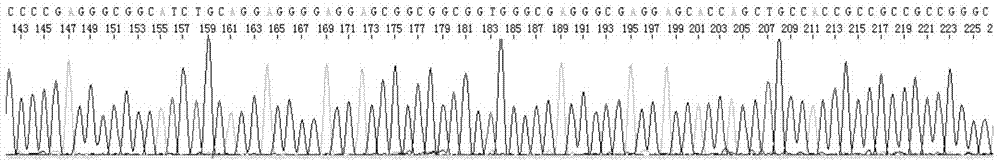
[0029] Example 1 Sequencing case of hairpin DNA sequence (ShDNA sequence, hairpin structure) using the sequencing method of the present invention
[0030] 1. In this embodiment, the PCR reaction system for sequencing the hairpin DNA samples consists of the following:
[0031] 5×sequencing buffer: 2μl
[0032] Template (150ng / μl): 1μl
[0033] Primer (3.2μM): 2μl
[0034] Sanger sequencing reaction optimizer 1 μl (by volume: 1 part of Bigdye, 0.2 part of dGTP-BigDye, 0.2 part of 10 μM dGTP and 0.2 part of 10 μM dCTP)
[0035] Water: 4 μl.
[0036] 2. The PCR reaction procedure for sequencing the hairpin DNA samples in this embodiment is as follows:
[0037] Pre-denaturation at 98°C for 8 minutes; quenching in ice water;
[0038] Take 98°C for 10 seconds, 50°C for 1 minute, and 60°C for 4 minutes as a cycle, and perform 40 cycles;
[0039] Store at 4°C.
[0040] 3. The steps of carrying out ethanol / sodium acetate purification to the PCR reaction product:
[0041] (1) Add...
Embodiment 3
[0070] Example 3 Sequencing case of sequences with high GC content using the sequencing method of the present invention
[0071] 1. In this embodiment, the PCR reaction system for sequencing samples with high GC content is composed of:
[0072] 5×sequencing buffer: 2μl
[0073] Template (150ng / μl): 1μl
[0074] Primer (3.2μM): 2μl
[0075] Sanger sequencing reaction optimizer 1 μl (by volume: 1 part of Bigdye, 0.5 part of dGTP-BigDye, 0.5 part of 10 μM dGTP and 0.5 part of 10 μM dCTP)
[0076] Water: 4 μl.
[0077] 2. The PCR reaction procedure for sequencing the samples with high GC content in this embodiment is:
[0078] Pre-denaturation at 98°C for 8 minutes; quenching in ice water;
[0079] Take 98°C for 10 seconds, 50°C for 1 minute, and 60°C for 4 minutes as a cycle, and perform 40 cycles;
[0080] Store at 4°C.
[0081] 3. Steps for purifying the PCR reaction product:
[0082] (1) Add 1 μl 125mM EDTA to the bottom of each tube, and add 1 μl 3M NaAc to the bottom...
Embodiment 4
[0089] Example 4. Comparative test example: Sequencing comparison of sequences with high GC content using conventional sequencing methods
[0090] This example is a comparative test example of Example 3, that is, sequences with high GC content are sequenced by conventional methods.
[0091] 1. PCR reaction system using conventional methods:
[0092] 5×sequencing buffer: 2μl
[0093] Template (150ng / μl): 1μl
[0094] Primer (3.2μM): 2μl
[0095] BDT: 1 μl
[0096] Water: 4 μl
[0097] 2. PCR program using conventional methods:
[0098] 95°C for 5 minutes; quenching;
[0099] 40 cycles were performed with 95° C. for 10 seconds, 50° C. for 1 minute, and 60° C. for 4 minutes as one cycle.
[0100] Store at 4°C.
[0101] 3. Steps for purifying the PCR reaction product:
[0102] (1) Add 1 μl 125mM EDTA to the bottom of each tube, and add 1 μl 3M NaAc to the bottom of each tube;
[0103] (2) Add 25 μl of 100% alcohol to each tube, mix well, and let stand at room temperature...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com