Hardware acceleration method for predication of RNA second-stage structure with pseudoknot
A secondary structure, hardware acceleration technology, applied in special data processing applications, instruments, electrical digital data processing, etc., can solve problems such as high space-time complexity, no hardware acceleration of high-dimensional four-dimensional structure prediction algorithms, and poor prediction results.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0050] The present invention will be further described below in conjunction with embodiment:
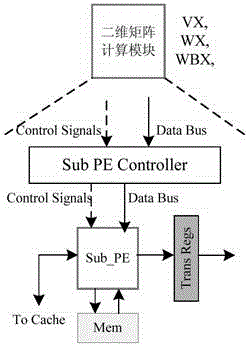
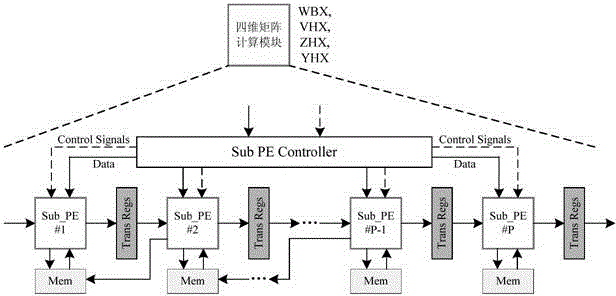
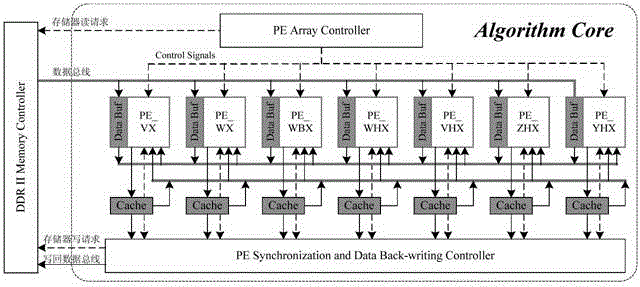
[0051] The present invention first constructs a heterogeneous computing system composed of a host and a reconfigurable algorithm accelerator, and then the host sends the formatted thermodynamic model parameters and encoded RNA sequences to the reconfigurable algorithm accelerator, and the seven computing modules of the algorithm accelerator The MPMD method is used to perform PKNOTS algorithm calculation without backtracking; in the calculation, the matrix dimensionality reduction method is used to decompose the four-dimensional matrix into N three-dimensional matrices, and then the task division strategy of layer-by-layer division by region and parallel processing by column within the region is used to achieve fine-grained Parallel computing, n PEs inside each computing module use SPMD to simultaneously calculate n data in different columns in the area, where n is a natural number.
...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com