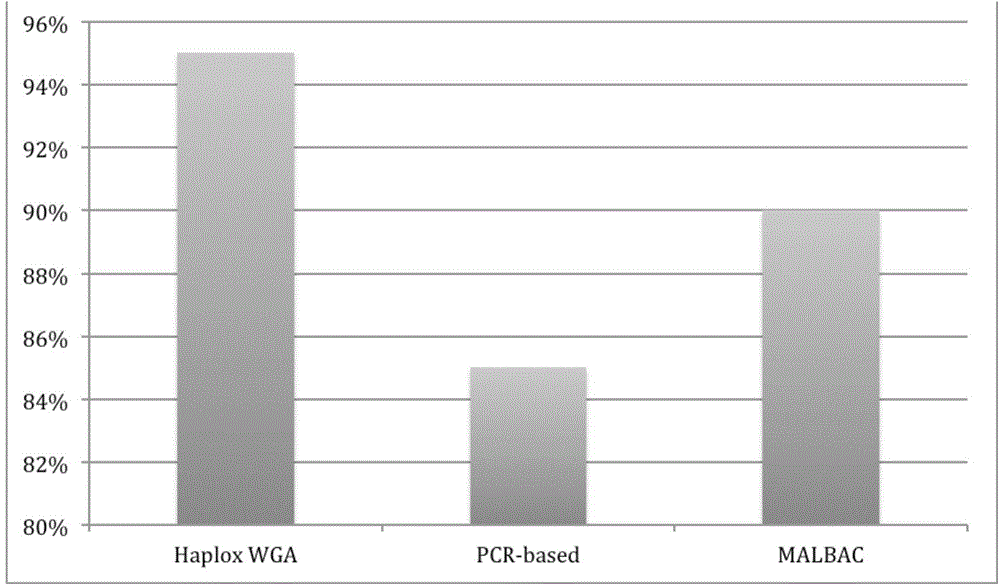
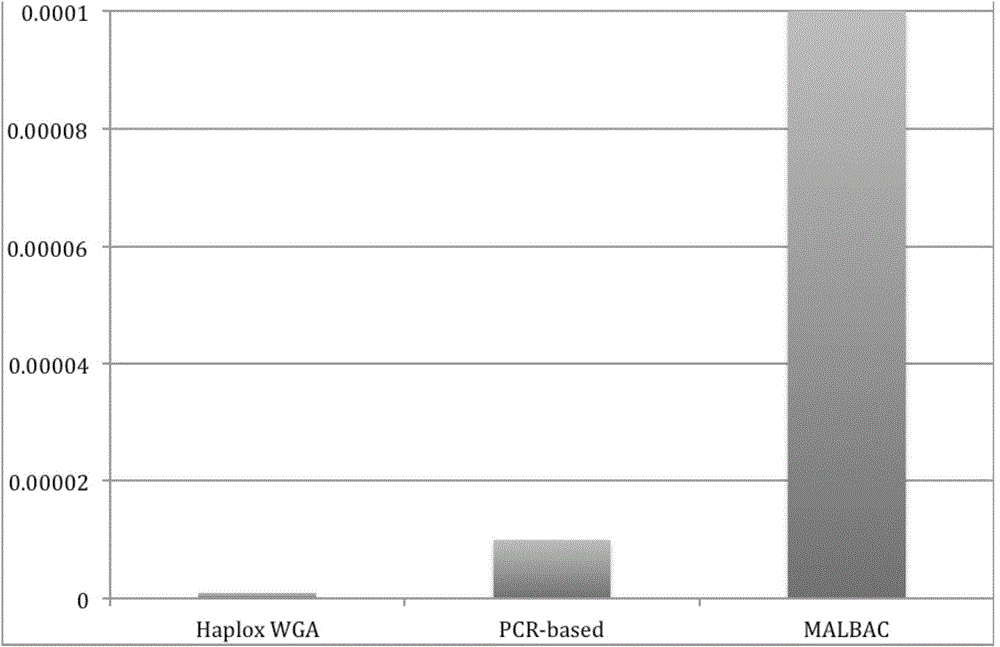
MDA (Multiple Displacement Amplification)-based whole-genome amplification method
A whole-genome amplification and genomic technology, applied in DNA preparation, recombinant DNA technology, etc., can solve problems such as errors, application limitations, and efficiency deviations, and achieve the effects of facilitating clinical testing, overcoming low coverage, and reducing error rates
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0029] A method for MDA-based whole-genome amplification, comprising the steps of,
[0030] 1) Separating and extracting a single tumor cell;
[0031] 2) extracting the genomic DNA of the single tumor cell by the method of Cell Search;
[0032] 3) Using the single-cell genomic DNA obtained in step 2) as a template, perform multiple displacement amplification (MDA) with the Haplox method single-cell whole-genome amplification kit, specifically, including the following steps:
[0033] Prepare mixed solution A: add random primer octamer and genomic DNA template to the amplification system, and the concentration of random primer octamer is 40 μM to obtain mixed solution A;
[0034] Prepare mixed solution B: freshly mix 10x Phi29Buffer, 1mM dNTPs, 0.1mM dUTP, H2O, and Phi29Polymerase (10U / μL, Enzymatics) to obtain mixed solution B; store at 4°C for later use;
[0035] Reaction: mix equal volumes of mixed solution A and mixed solution B into the reaction tube, then add mineral oil...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com