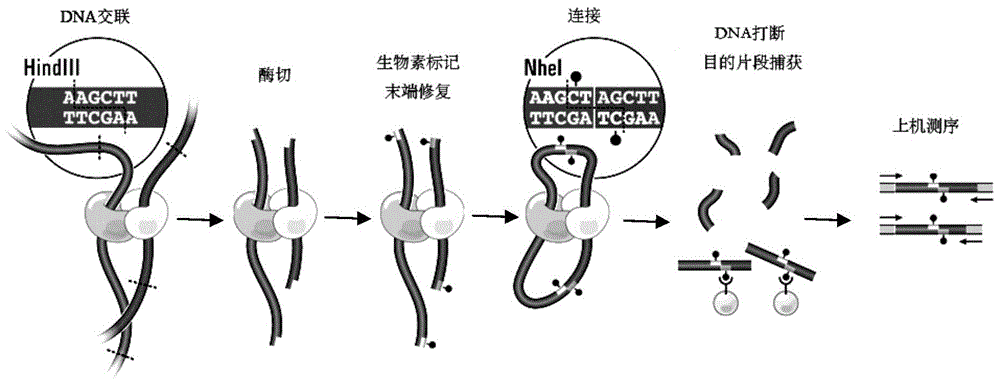
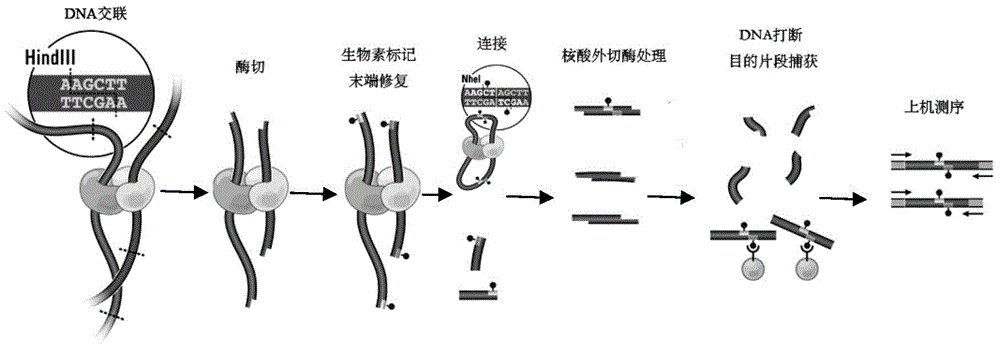
High-throughput sequencing library and its construction method
A construction method and technology for sequencing libraries, applied in libraries, chemical libraries, nucleotide libraries, etc., can solve the problems of low sequencing efficiency and small amount of effective data, and achieve the effects of reducing invalid data, improving purity, and increasing effective amount
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0036] It should be noted that, in the case of no conflict, the embodiments in the present application and the features in the embodiments can be combined with each other. The present invention will be described in detail below with reference to the accompanying drawings and examples.
[0037] Exonuclease in the present invention refers to the nuclease that acts on double-stranded DNA, has 3 ' → 5 ' exonuclease activity but does not have DNA polymerase activity. What the exonucleases applicable to the present invention have in common is that they all use double-stranded DNA as a substrate, and have the activity of excising single nucleotides one by one from the 3'-OH end without the activity of DNA polymerase , Nucleases that meet the above conditions are all within the protection scope of the present invention, and, in addition to having the above-mentioned activities, the exonucleases of the present invention can also have other relatively weak nuclease activities, such as B...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com