Small interfering RNA targeting human ran and its use in the preparation of anti-hepatitis C medicaments
A small interference, drug technology, applied in the field of medicine, can solve the problem of not being able to block the transmission of HCV
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
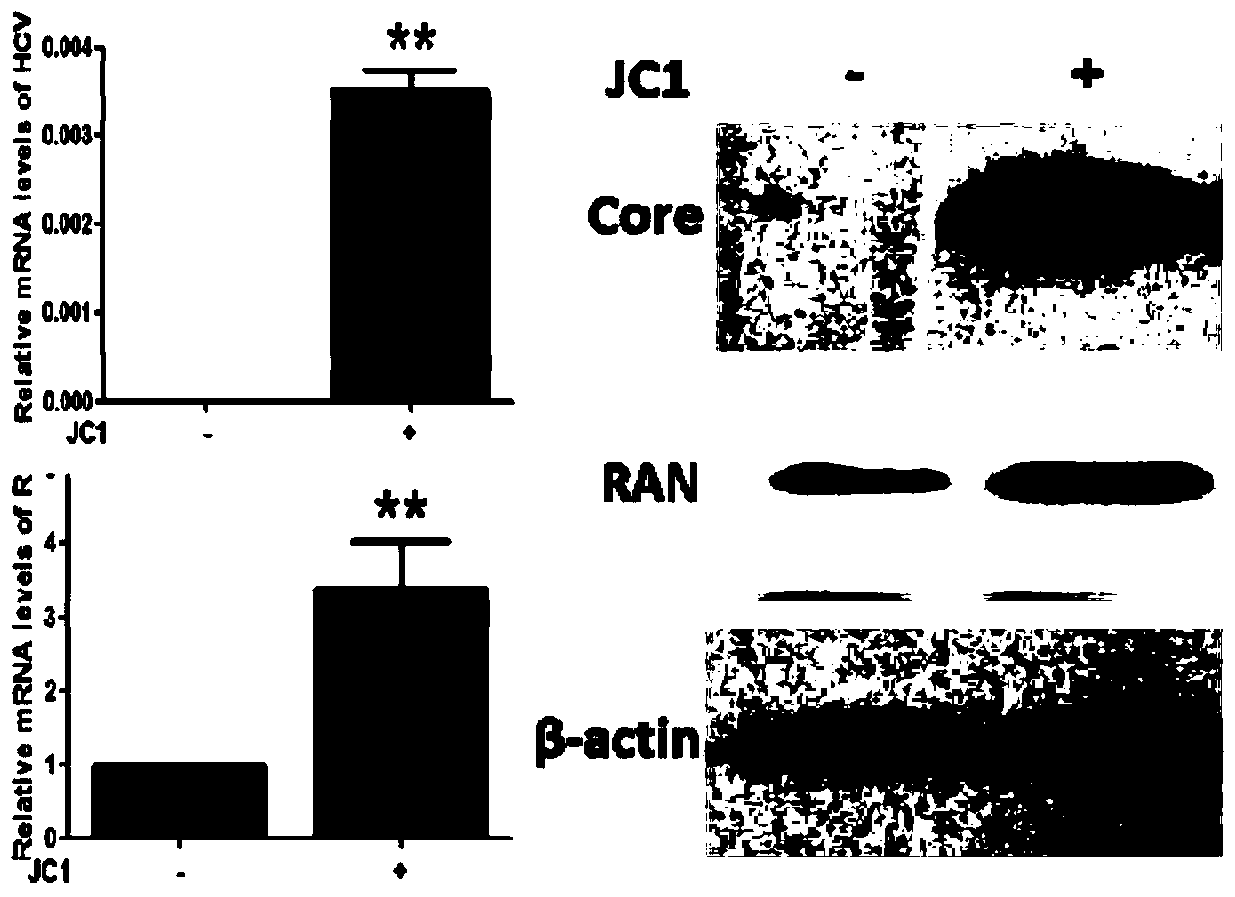
[0026] Example 1 Analysis of RAN expression level after JC1 infection
[0027] Establishment of HCV Cell Infection Model
[0028] 1 day before transfection, with 1×10 7 Inoculate a 10 cm culture dish at a density of cells / well, and the degree of cell confluence is 70% to 80%. During transfection, 10 μg of HCV RNA transcripts were introduced into the liver cancer cell line Huh7.5.1 supporting HCV replication by electrotransfection with a single square wave at 260V and a pulse length of 25 milliseconds. The cell supernatant was collected, centrifuged at 1500 g for 5 min to remove cell debris, and stored at -70°C. Replant Huh7.5.1 cells in a six-well cell culture plate. When the cells reach 50% to 60% confluence, add 2ml of the collected cell supernatant to each well, incubate for 24 hours, wash with PBS three times, and add complete medium to continue culturing . Cells were harvested 48 hours later for subsequent experiments.
[0029] Fluorescent quantitative PCR
[0030] ...
Embodiment 2
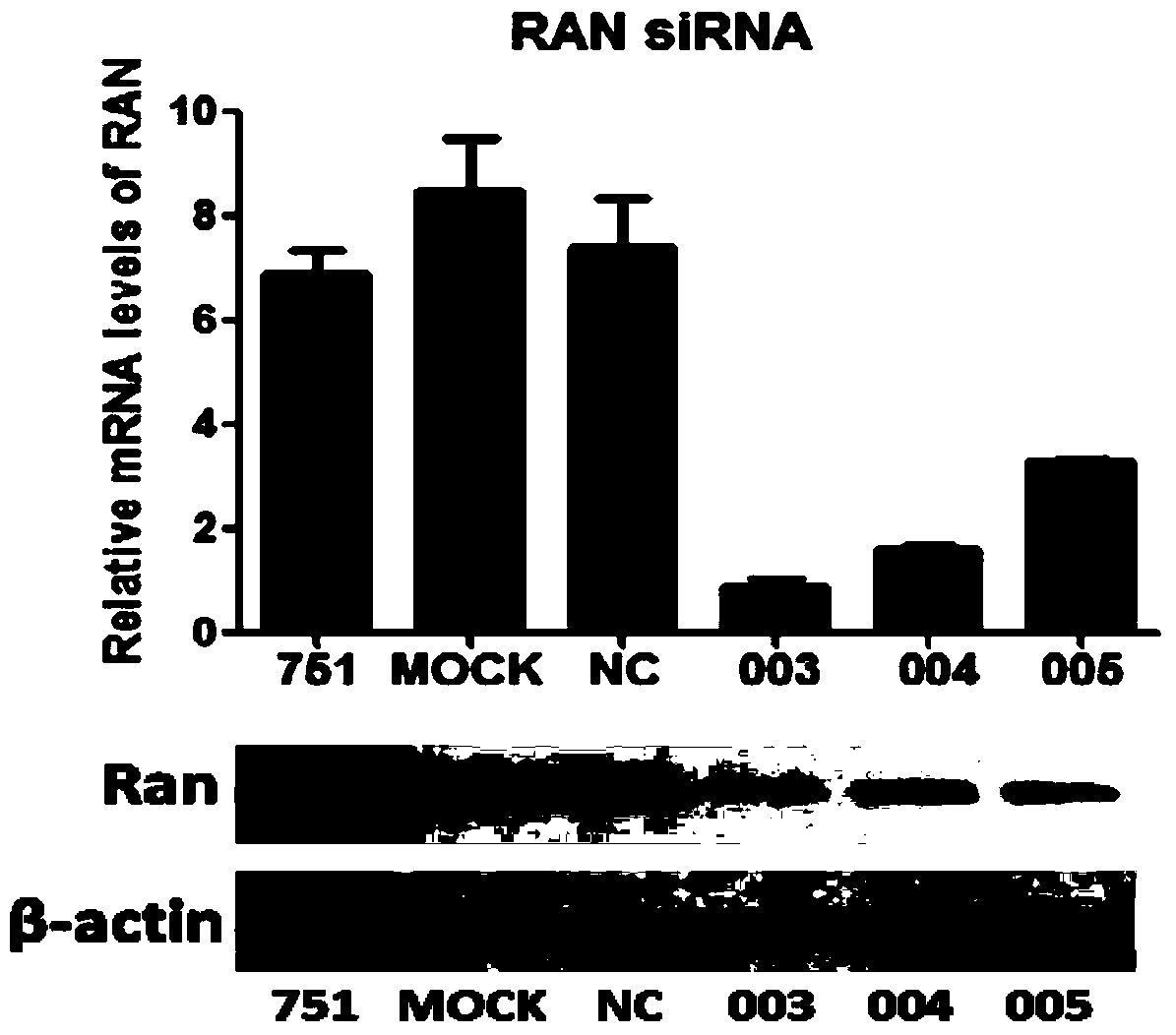
[0034] Example 2 Screening of RAN interference sequences
[0035] RNA interference
[0036] 1 day before transfection, with 2×10 5 6-well cell culture plates were inoculated at a density of cells / well. For transfection, take 10 μl siRNA (commissioned by Ribo Biosynthesis) and dilute it with 100 μl Opti-MEM I serum-free medium, mix gently; take 2 μl Lipofectamine TM For RNAiMAX, dilute in 100μl Opti-MEM I serum-free medium and mix gently. Diluted HCV RNA and Lipofectamine TM RNAiMAX was mixed gently and allowed to stand at room temperature for 20 minutes. Add 200 μl of the complex to the cells in the 6-well plate and mix gently. After 24 hours, add 2ml of JC1 virus supernatant to each well to infect the cells, and after incubation for 24 hours, add complete medium to continue culturing. Cells were harvested 48 hours later for subsequent experiments.
[0037] Fluorescent quantitative PCR
[0038] Fluorescent quantitative PCR was used to detect the relative amount of HCV ...
Embodiment 3
[0042] Embodiment 3 003 anti-HCV effect analysis
[0043] Western Blot detection
[0044] The expression level of HCV core protein was detected by Western Blot. After RNA interference, the total cellular protein was extracted and quantitatively analyzed according to the operating instructions of the BCA protein quantification kit. 40 μg of the above-mentioned total cellular protein was separated by SDS-PAGE and transferred to a nitrocellulose membrane. After blocking with 5% skimmed milk powder, the membrane was incubated with antibodies HCV Core (Abcam) and β-actin (Cellsignaling Technology) at 4°C overnight, and the secondary antibody was incubated for 1 hour and detected with chemiluminescence reagent (Pierce).
[0045] Such as image 3 As shown, 003 can significantly inhibit the expression of HCV core protein.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com