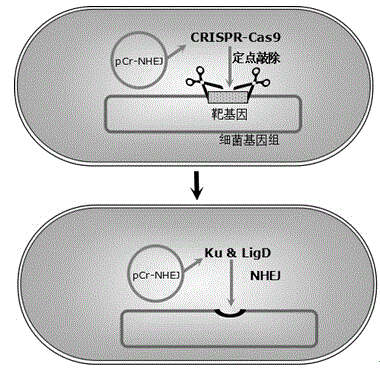
PCr-NHEJ (non-homologous end joining) carrier as well as construction method of pCr-NHEJ carrier and application of pCr-NHEJ carrier in site-specific knockout of bacterial genes
A construction method and a technology for targeted knockout, applied in the field of genetic engineering, can solve the problems of cumbersome operation, difficult operation, and insufficient gene knockout efficiency, and achieve the effects of low operation difficulty, reduced operation difficulty, and high gene knockout efficiency.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
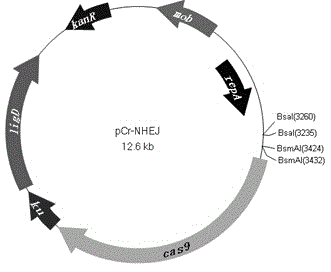
[0061] A pCr-NHEJ vector, the sequence of the pCr-NHEJ vector has the sequence shown in SEQ ID NO:1. The pCr-NHEJ vector of this embodiment can be used for bacterial gene knockout, and has the advantage of high gene knockout efficiency.
Embodiment 2
[0063] The application of a pCr-NHEJ vector of Example 1 in site-specific knockout of bacterial genes.
Embodiment 3
[0065] The construction method of a pCr-NHEJ vector of Example 1, includes the following steps:
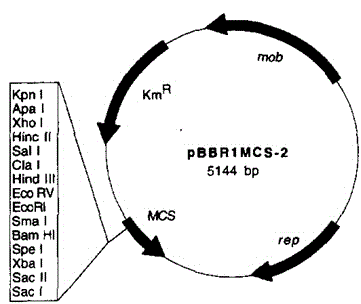
[0066] (1) Use broad host plasmid of gram-negative bacteria P BBR1MCS-2 (GI: 773412) as the basic carrier; cas9
[0067] (2) The following three protein genes were amplified using the upstream primer and downstream primer with the prokaryotic promoter PLtetO-1 for mRNA transcription: cas9 (GI:674296984), ku (GI:444893469) and ligD (GI:444893469), respectively cas9 Amplification products, ku Amplification products and ligD Amplification product; among them, cas9 , ku with ligD The open reading frames of the three protein genes are all located downstream of independent PLtetO-1, so that cas9 , ku with ligD The expression of the three protein genes is regulated by the promoter; cas9 Amplification products, ku Amplification products and ligD There is a 15nt base overlap between the amplified products;
[0068] (3) cas9 The 3’ end of the amplified product and ku The 5'end of the amplified...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com