Cas9 gene knockout vector suitable for endophytic fungi A761 of Morinda officinalis, construction method and application thereof
An endophytic fungus and construction method technology, applied in the fields of biochemistry and molecular biology, can solve the problem of fewer filamentous fungi, achieve convenient construction, high gene knockout efficiency, and promote development and utilization
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
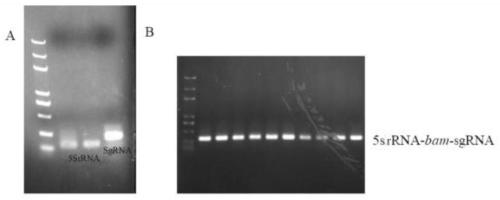
[0021] Construction of Example 1 Gene Knockout Vector PFC332-sgRNA-bam
[0022] According to the BAM sequence, the BAM target sequence 5'-GCAGTGGGAACAGGAGAGAT-3' was designed on the website http: / / www.e-crisp.org / E-CRISP / index.html. Design the 5srRNA promoter, the sgRNA containing the target sequence and the corresponding terminator fragment, and integrate the two fragments together by homologous recombination to obtain the 5SrRNA-bam-sgRNA-ter fragment, whose sequence is shown in SEQ ID NO.1. details as follows:
[0023] Design 5SrRNA amplification primers, respectively 5SrRNA-F: 5′-CGGGAAGATCTCACATACGACCACAGGG-3′, 5SrRNA-R: 5′-CATACAACAGAAGGGATTCGCTGGTG-3′,
[0024] Amplification primers of the sgRNA fragment containing the target sequence: sgRNA-F: 5′-αCGAATCCCTTCTGTTGTGCAGTGGGAACAGGAGAGATGGGGTTTTTAGAGCTAGA-3′, sgRNA-R: 5′-GTCTTAATTAAGCGGCCCCTCTAGATGCATGC-3′.
[0025] Primers were synthesized by Guangzhou Tianyi Huiyuan Company.
[0026] Using the 5SrRNA promoter fragmen...
Embodiment 2
[0031] Knockdown of the cytosporaphenones biosynthesis gene bam in C. rhizophorae A761:
[0032] The method for introducing the recombinant knockout vector PFC332-sgRNA-bam into C. rhizophorae A761 protoplasts is as follows:
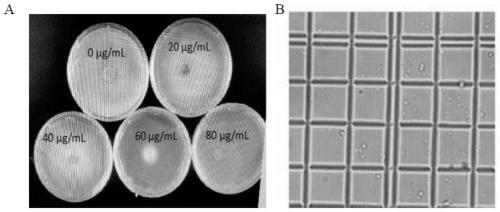
[0033] The C. rhizophorae A761 hyphae were transferred to PDA plates with hygromycin concentrations of 0 μg / mL, 20 μg / mL, 40 μg / mL, 60 μg / mL, and 80 μg / mL for hygromycin resistance screening. The results showed that C.rhizophorae A761 could not grow when the hygromycin concentration was 80 μg / mL ( image 3 A).
[0034] Cytospora rhizophorae A761 mycelia were inoculated in PDA liquid medium and cultured for 7 days, 2 g of the better-growing bacteria were taken, and the bacteria were washed twice with PBS. Dissolve 0.15 g of lyase in 20 mL of KC buffer, filter with a 0.22 μm filter membrane, then add the washed bacteria, and lyse at 28°C for 3 hours at 68 r / min. Pour the lysed bacterial solution into a strainer to filter to remove mycelium, and filter a...
Embodiment 3
[0037] Knockout verification and product comparison analysis of the bam gene in Example 3C.rhizophorae A761
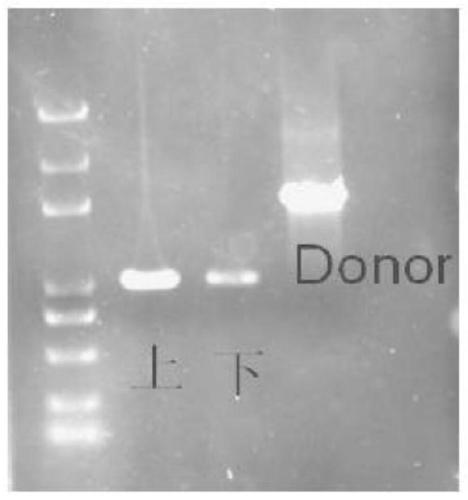
[0038] Genomic DNA was extracted from the wild C. rhizophorae A761 strain and the bam knockout C. rhizophorae A761 strain, using the left homology arm upstream primer LF and the right homology arm primer RR as primers, using the extracted genome as a template, and using Prime STAR MAX (TAKARA, Japan) for amplification. The results showed that when the genomic DNA of the wild A761 strain was used as a template, a target fragment of 3.5 kb could be amplified, and when the genomic DNA of the bam gene knockout C. rhizophorae A761 strain was used as a template, a fragment of about 1.5 kb was obtained, proving that bam The gene was successfully knocked out ( Figure 5 ).
[0039] Inoculate the wild C.rhizophorae A761 strain and the bam gene knockout C.rhizophorae A761 strain into PDA medium, and culture them at 28°C and 180rpm for 7 days. The fermentation product was extr...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com