Method for identifying copy number of T-DAN tandem repeat sequences in transgenic plant through real-time fluorescence quantification PCR method
A real-time fluorescence quantitative, tandem repeat sequence technology, applied in recombinant DNA technology, plant products, genetic engineering, etc., can solve the problem that the number of copies of T-DNA cannot be determined, and the formation mechanism of the tandem repeat structure of T-DNA is not completely clear.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0025] The present invention will be described in further detail below according to the drawings and embodiments.
[0026] A method for identifying the copy number of T-DNA tandem repeat sequences in transgenic plants by real-time fluorescent quantitative PCR method, the steps comprising:
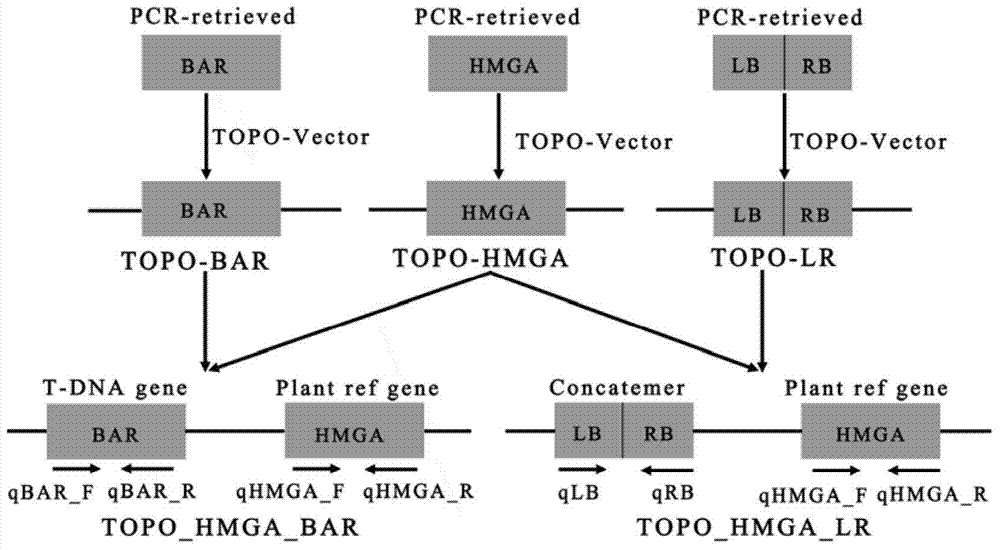
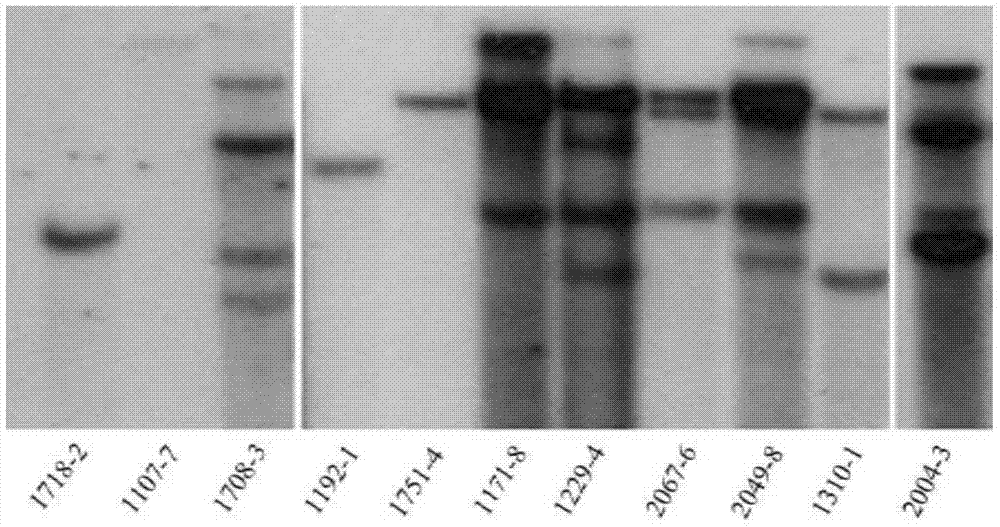
[0027] Through the Agrobacterium-mediated plant transformation method, the plant expression vector pSKI015 containing the screening gene BAR was connected to the HMGA gene, and transformed into wild-type Col-4 Arabidopsis thaliana, and 300 independent transgenic lines were obtained;
[0028] Use two reference plasmids such as the sequence listing, the two reference plasmids all contain an internal reference gene encoding Arabidopsis high mobility protein HMGA, one of the plasmids is to connect the HMGA gene to the BAR gene on the PSKI015 vector, and the other plasmid is Connect the HMGA gene with the T-DNA direct repeat sequence containing the left border LB and the right border RB to form ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com