Method of carrying out rapid and efficient DNA combination and evolution based on synthesis of single-chain DNA library
A single-chain, high-efficiency technology that can be used in the field of genetic engineering to solve the problem of wasting cell synthesis capacity and the burden of cell growth.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
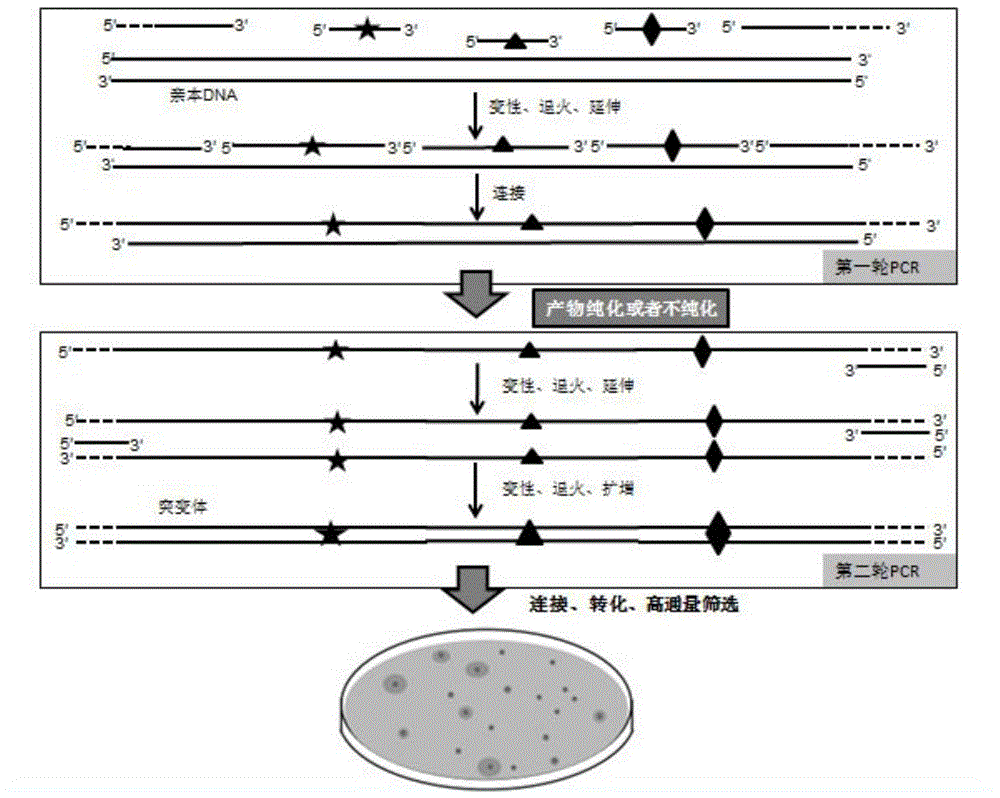
[0049] Embodiment 1 RECODE technology two-round PCR reaction system prepares combinatorial mutation library
[0050] A. Preparation of double-stranded mutant library after purification of single mutant library
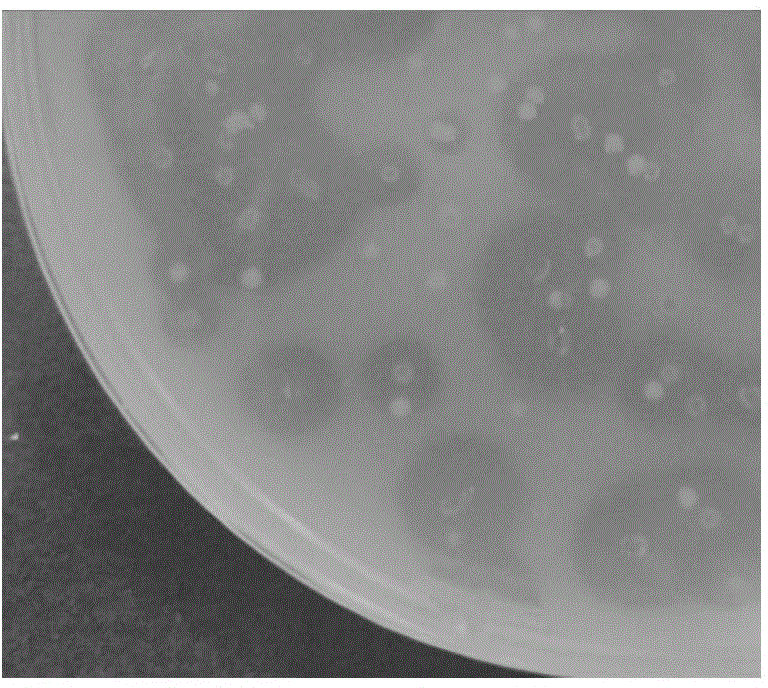
[0051] Take the genes of β-galactosidase and esterase lacZ-Esterase as an example to carry out combined lethal mutations, and the nucleotide sequence is shown in SEQ ID NO.1. In this example, galactosidase and esterase genes are used for combined lethal mutation, and the diversity of the library can be indicated by plate screening. Substrates 0.1 mM IPTG, 20 μg / ml X-Gal and 1% Tris Glyceryl Butyrate. The two genes were recombined into the pMD19 vector and transformed into the JM109 host. On the screening plate, the parental gene host such as image 3 As shown, the colony is blue (lacZ degrades X-gal), and produces a hydrolysis transparent circle (Esterase hydrolyzes tributyrin).
[0052] β-galactosidase (with promoter) downstream tandem esterase gene, carry out comb...
Embodiment 2
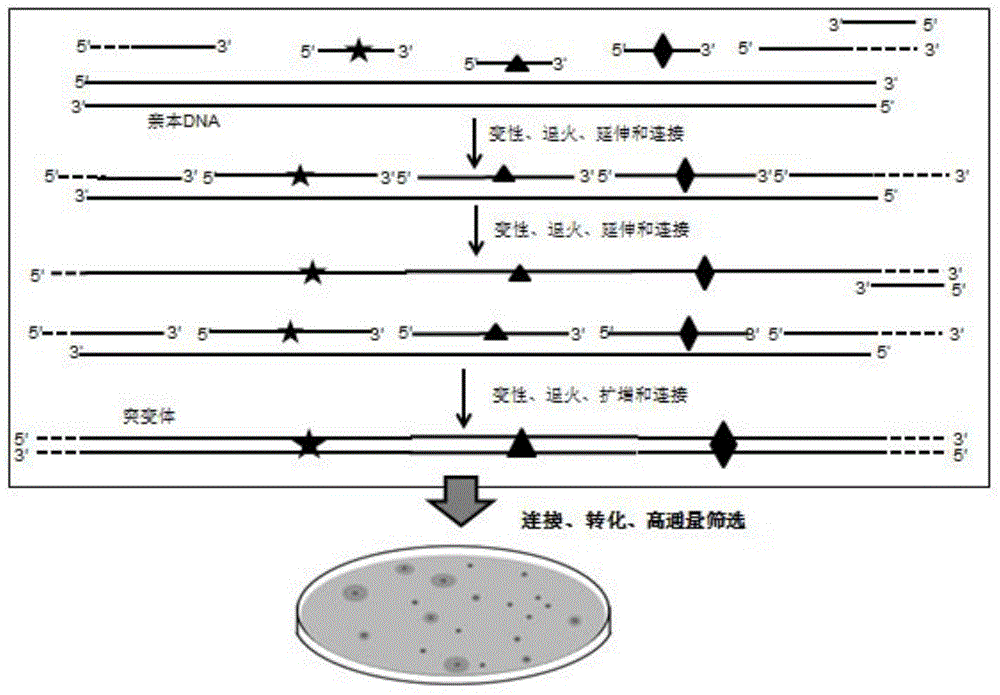
[0066] Embodiment 2 RECODE technology one-step PCR reaction system prepares combinatorial mutation library
[0067] In this example, in order to further simplify the RECODE technique, the genes of β-galactosidase and esterase lacZ-Esterase in Example 1 were also used as an example to carry out combined lethal mutations. Editing primers JPS-MD / LacZ-P1F, JPS-MD / Est-P2F, JPS-MD / Est-P3F and downstream anchor primer JPS-LZE / DTA were mixed in equimolar ratio, and phosphorylated by T4 polynucleotide kinase For chemical reaction, the reaction system was operated according to the product instructions, reacted at 37°C for 30 minutes, and inactivated the enzyme at 70°C for 10 minutes. One-step reaction to prepare a double-stranded mutant library is a simultaneous reaction of PCR cycles for DNA extension, ligation, and amplification of mutant strands.
[0068] In a 50 μl reaction system: 5 μl 10x reaction buffer, upstream anchor primer JPS-LZE / UTA, phosphorylated downstream anchor primer...
Embodiment 3
[0071] Example 3 In vitro transformation of the constitutive promoter of the rpoS gene of Escherichia coli
[0072] Taking the constitutive promoter of the rpoS gene of Escherichia coli as an example, the nucleotide sequence is shown in SEQ ID NO.2, and the four internal spacer sequences in the -10 and -35 intervals were combined and mutated simultaneously to construct a mutant library. Four editing primers were designed according to the sequence of the promoter segment, editing primers (Jps / rpoSp-F, Jps / rpoSp4-F, Jps / rpoSp3-F, Jps / rpoSp21-F), anchor primers (Jps / rpoSp-HM -F, Jps / rpoSp-HM-R) outer primers (rpoSp-F, rpoSp-R) and vector preparation primers are as follows:
[0073] Jps / rpoSp-F:TTCCACCGTTGCTGTTGCGTNNNNNNNNNNNNNNNNTATTCTGAGTCTTCGGGTGAAC
[0074] Jps / rpoSp4-F:CATAACGACACAATGCTGGTNNNNNNNNNNNNNAAGTTAAGGCGGGGCAAAAAATAGC
[0075] Jps / rpoSp3-F:TAGCACCGGAACCAGTTCAANNNNNNNNNNNNNNNNAATTCGTTACAAGGGGAAATCCG
[0076] Jps / rpoSp21-F:GCAGCGATAAATCGGCGGAACNNNNNNNNNNNNNNNNNTGNTC...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com