Rapid Bacterial Resistance Prediction System and Its Prediction Method
A technology for predicting system and drug resistance, applied in the field of public health and environment, which can solve the problems of isolated measurement data, inconvenient viewing, retrospective data, and inability to establish long-term and effective epidemic files of drug-resistant strains, so as to eliminate differences and reduce costs. speed effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0045] The present invention will be described in further detail below with reference to the embodiments and the accompanying drawings, but the embodiments of the present invention are not limited thereto.
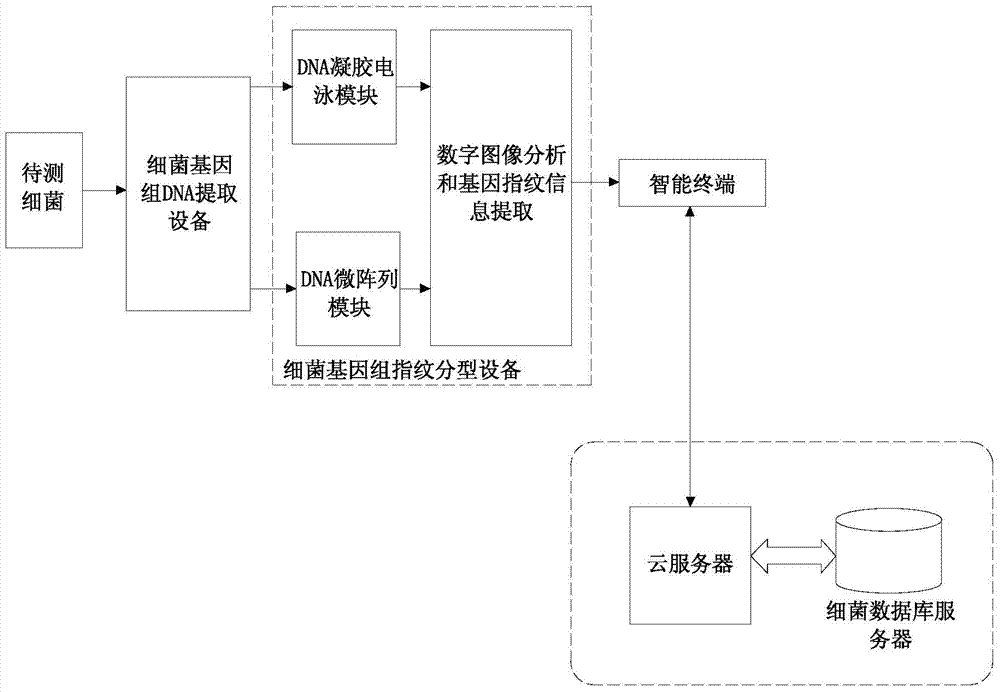
[0046] like figure 1 As shown, the bacterial drug resistance rapid prediction system of this embodiment includes bacterial genomic DNA extraction equipment, bacterial genomic fingerprint typing equipment, intelligent terminal, cloud server and bacterial database server. Type equipment is connected, and the bacterial genome fingerprint typing equipment is connected through lines (such as USB, serial port UART, bus SPI, I 2 C, etc.) or wireless signals (such as WIFI, Bluetooth, etc.) are connected to the intelligent terminal, and the intelligent terminal is connected to the Internet through the Internet (wired or wireless, such as Ethernet, cable modem, WIFI, Bluetooth, GSM / GPRS, 3G, 4GLTE, etc.) is connected with a cloud server, and the cloud server is encrypted and connecte...
Embodiment 2
[0120] like figure 1 and Figure 5 As shown, the software flow of the bacterial drug resistance rapid prediction system of this embodiment is implemented through the following steps:
[0121] A. The user will obtain a sample of bacterial genomic DNA to be tested in the bacterial genomic DNA extraction equipment;
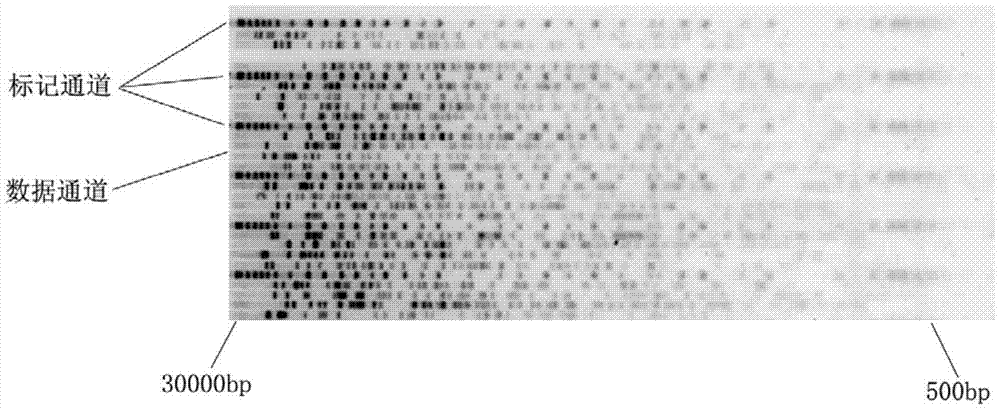
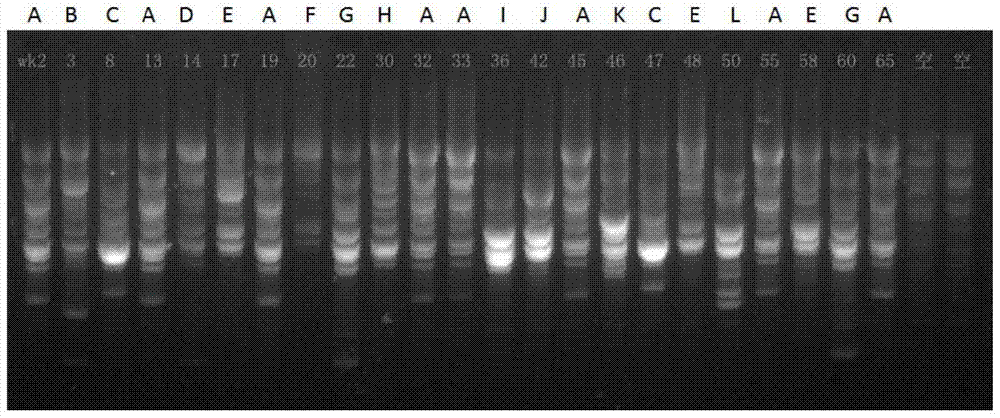
[0122] B. The user places the DNA sample in the gel electrophoresis module of the bacterial genome fingerprinting equipment to generate a striped fingerprint and the signal is collected by the image collector;
[0123] C. The user places the DNA sample in the DNA microarray module of the bacterial genome fingerprinting equipment to generate a microarray-like fingerprint and the signal is collected by the image collector;
[0124] D. The user sends the DNA collected by the bacterial genome fingerprinting equipment through the line (such as USB, serial port UART, bus SPI, I 2 C, etc.) or wireless signals (such as WIFI, Bluetooth, etc.) are sent to the smart terminal...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com