Molecular marker method for rapidly identifying suitability of tea germplasms for manufacture of oolong
A technology of molecular markers and oolong tea, which is applied in the direction of biochemical equipment and methods, microbiological determination/inspection, etc., can solve the problems of long identification time, labor-consuming, low identification efficiency, etc., to shorten the identification cycle, reduce manpower, and reduce blindness sexual effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
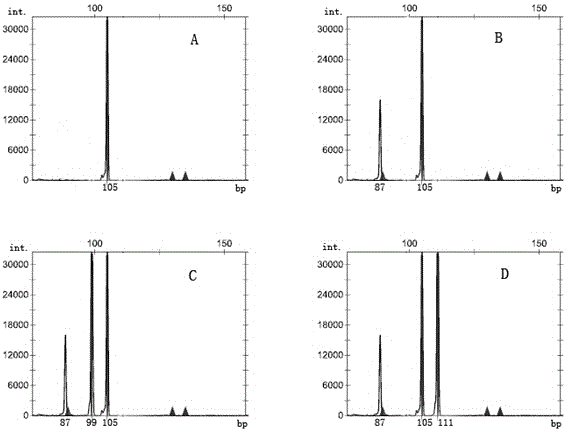
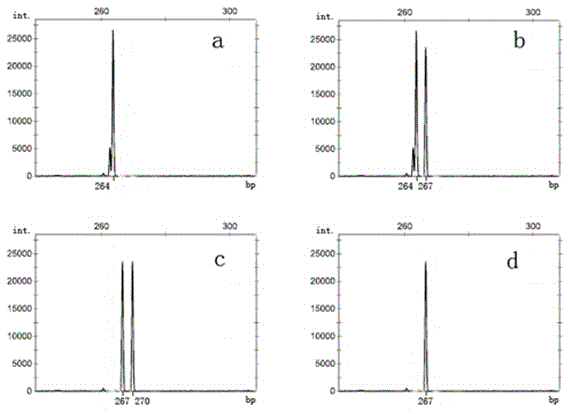
Image
Examples
Embodiment 1
[0058] Example 1. Determination of the new tea tree strain Chuntaoxiang (strain number 115)
[0059] Chuntaoxiang is a new strain of tea tree clones bred by the Tea Research Institute of Fujian Academy of Agricultural Sciences by hybrid breeding method. Both the strain comparison test and the regional adaptability test show that this strain is suitable for oolong tea ([1] Guo Jichun, et al. Comparative identification of oolong tea strains and selection. Tea Science Briefing, 1994, (145) 4:13-21. [2] Guo Jichun, et al. Preliminary report on regional trials of new varieties of oolong tea. Tea Science and Technology, 1995,149(4):15-20).
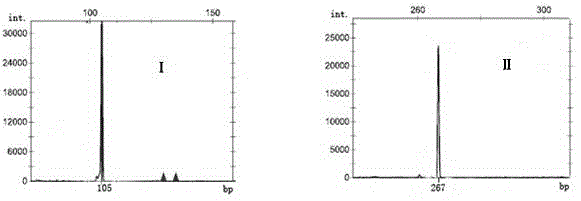
[0060] According to the above steps (1) to (4), the genotypes of the new tea tree line Chuntaoxiang at the TM352 and TM445 loci were respectively obtained ( image 3 , Ⅰ, Ⅱ), and then compare it with the genotype of the tea tree germplasm suitable for making oolong tea at the TM352 and TM445 sites, and the genotype is compared with the tea tre...
Embodiment 2
[0061] Example 2, Determination of new tea strain Mingke No. 3 (strain number 511)
[0062] Minke No. 3 is a new strain of tea tree clone bred by the Tea Research Institute of Fujian Academy of Agricultural Sciences by hybrid breeding method. Regional adaptability tests show that this strain is suitable for red and green tea, but not suitable for oolong tea (Zhou Fuyu. Tea tree hybrid Mingke 3 No. National Regional Test Guizhou Point Test Report. Fujian Tea, 2009, 4:5-7).
[0063] According to the above steps (1) to (4), the genotypes of the new tea tree line Mingke No. 3 at the TM352 and TM445 loci were respectively obtained ( Figure 4 , Ⅰ, Ⅱ), and then compare it with the genotypes at the TM352 and TM445 sites of the tea tree germplasm suitable for making oolong tea, although the genotype at the TM445 site of the germplasm ( Figure 4 , Ⅱ) and the genotype of the tea germplasm suitable for oolong tea at the TM445 locus ( figure 2 , a) Consistent, but the genotype of th...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com